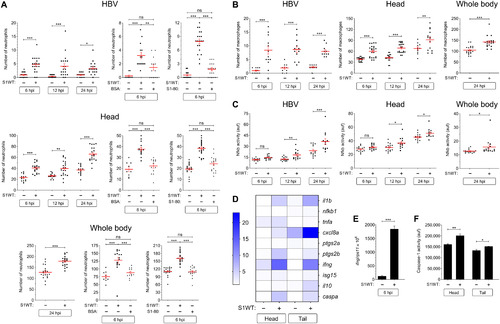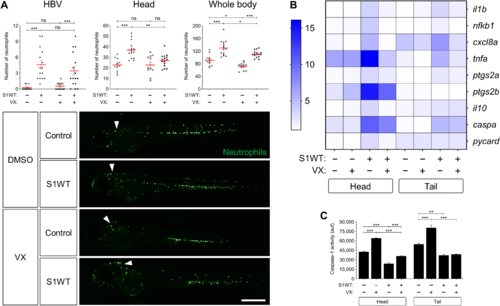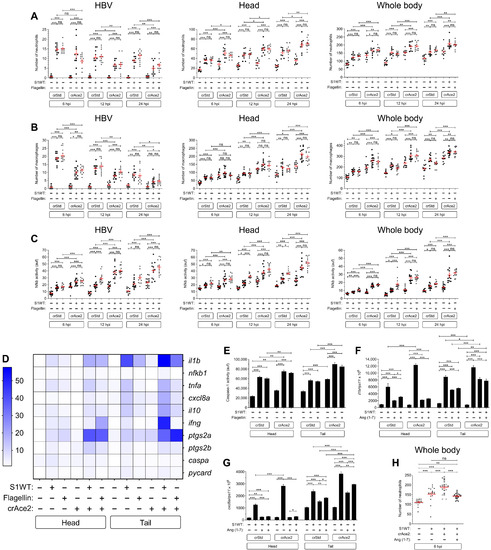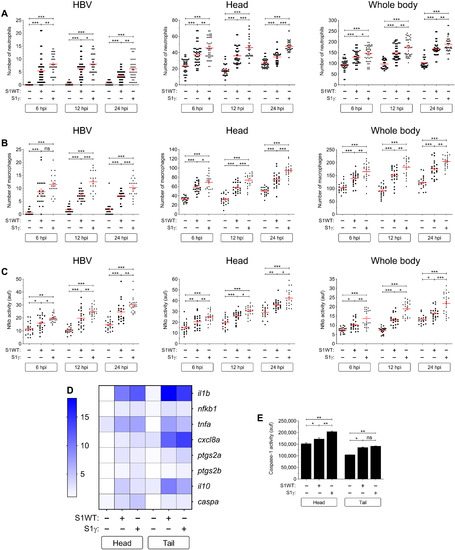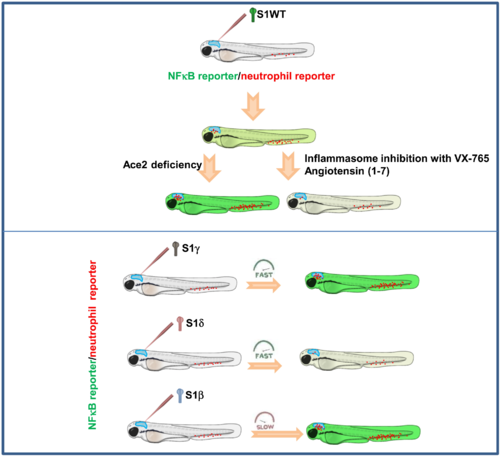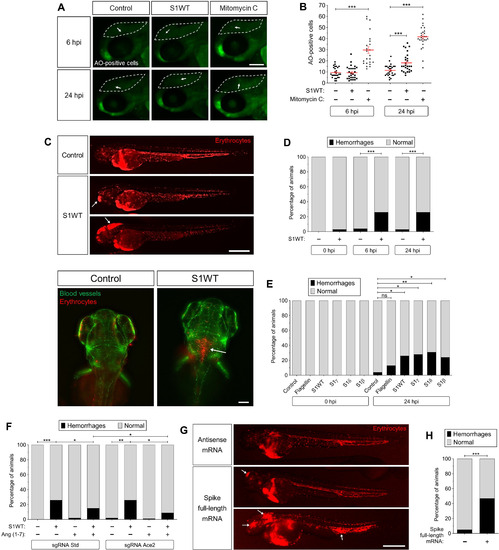
Fig. 7. Recombinant S1 proteins provoke hemorrhages in zebrafish larvae. (A and B) WT larvae were injected with S1WT protein, mitomycin C, or vehicle (−) in the hindbrain ventricle at 2 dpf and the number of cell death in the brain analyzed at 6 and 24 hpi by acridine orange (AO) staining. (C to F) One-cell-stage zebrafish eggs of Tg(gata1a:dsRed)/Tg(fli1a:eGFP) were microinjected with control or ace2 crRNA-Cas9 complexes. At 2 dpf, S1 proteins or flagellin were injected in the hindbrain ventricle alone or with Ang (1-7). Hemorrhages were analyzed at 0, 6, and 24 hpi. (G and H) Zebrafish eggs of Tg(gata1a:dsRED) were microinjected with control or full-length S RNAs, and hemorrhages were analyzed at 48 hpf. Representative images showing AO+ cells (A) and hemorrhages (white arrows) (C and G). P values were calculated using one-way ANOVA and Tukey multiple range test (B) and Fisher’s exact test (D, E, G, and H). (D) 0 hpi + H2O, n = 34; 0 hpi + S1WT, n = 50; 6 hpi + H2O, n = 50; 6 hpi + S1WT, n = 60; 24 hpi + H2O, n = 52; and 24 hpi + S1WT, n = 60. (E) 0 hpi + H2O, 35; 0 hpi + flagellin, n = 35; S1WT, n = 47; S1γ, n = 40; S1δ, n = 42; S1β, n = 46. (F) crSTD + H2O, n = 92; crSTD + S1WT, n = 85; crSTD + ANG (1-7), n = 83; crSTD + S1WT + ANG (1-7), n = 87; crAce2 + H2O, n = 76; crAce2 + S1WT, n = 83; crAce2 + ANG (1-7), n = 90; crAce2 + S1WT + ANG (1-7), n = 81. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. Scale bars, 100 μm (A), 500 μm (top of C, and G), 50 μm (bottom of C).
|