- Title
-
A complex genetic architecture in zebrafish relatives Danio quagga and D. kyathit underlies development of stripes and spots
- Authors
- McCluskey, B.M., Uji, S., Mancusi, J.L., Postlethwait, J.H., Parichy, D.M.
- Source
- Full text @ PLoS Genet.
|
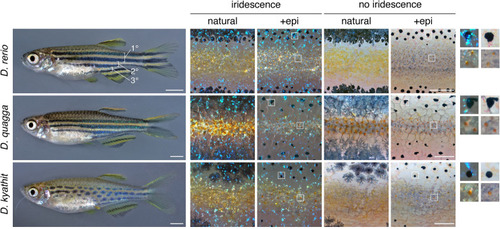
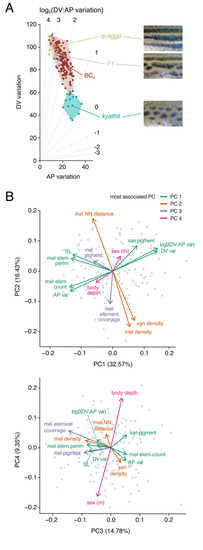
Top, Striped pattern of D. rerio showing primary (1°) light interstripe with primary dark stripes dorsally and ventrally. Secondary and tertiary interstripes and stripes are indicated only ventrally. Right, Cells that comprise dark and light pattern elements. Lighting has been adjusted to highlight or eliminate the iridescence of iridophores, and pigment cells are shown both in their natural state and after treatment with epinephrine (epi), which contracts pigment granules of melanophores and xanthophores towards cell centers. Insets at far right show higher magnification views of boxed regions. Dark stripes comprise melanophores with black melanin granules as well as more superficial iridophores that have a bluish iridescence owing to precisely oriented stacks of guanine-containing reflecting platelets (upper insets). Interstripes and interspot regions contain densely packed iridophores with a yellowish hue owing to disordered stacks of reflecting platelets, as well as more superficially located xanthophores, marked by lipid droplets containing yellow–orange carotenoids [6,54]. Middle and Bottom, Striped and spotted patterns of D. quagga and D. kyathit. Cell types present in each species were indistinguishable from those of D. rerio. In older D. kyathit and D. quagga, red pigment cells—erythrophores—were present and especially prominent in males, whereas D. quagga developed fissures and other disruptions in their stripes, especially in females (S1 Fig). Neither of these features occurs in D. rerio and we do not consider them further in this study. Scale bars, 2 mm (left) and 200 μm (right). |
|
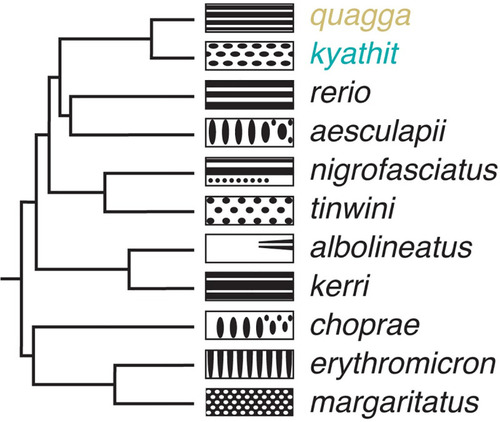
Phylogeny of Danio species, recovered in [16], with schematics illustrating adult melanophore pigment pattern variation (left is anterior). Danio quagga has also been dubbed D. aff. kyathit [16,19,47,49] and individuals likely representing the same species were misidentified originally as D. rerio [97]. Full color images of live fish can be found in [19] (open access); additional images can be found in [4,16]. |
|
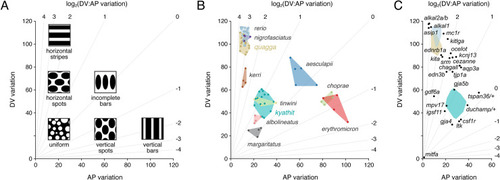
(A) Schematic illustrating where idealized patterns fall in a morphospace defined by DV (dorsoventral) and AP (anteroposterior) pattern variation of melanized elements. Diagonals indicate different log2 values for the ratio of DV variation: AP variation. (B) Mapping of pattern phenotypes in this morphospace revealed overlap of striped PHENOTYPE:
|
|
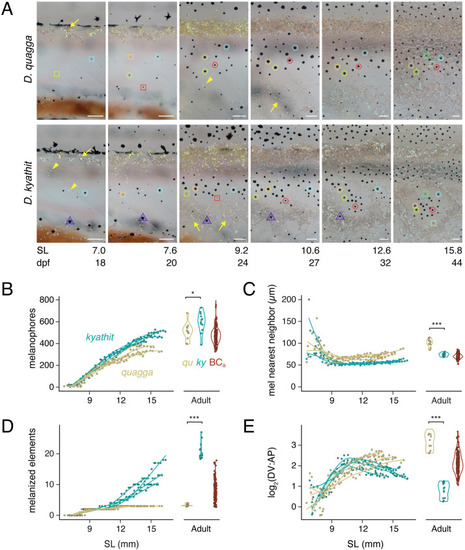
(A) Representative individuals imaged repeatedly during adult pattern formation. Images are aligned to show corresponding regions and rescaled to control for overall growth (frames and regions are selected from |
|
(A) Pattern variation morphospace showing locations of parental |
|
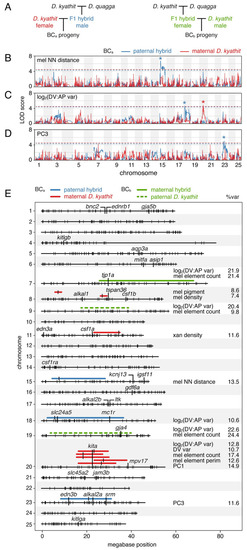
(A) Crossing schemes for mapping variation in two independent families, BCa and BCb (see main text). (B–C) Log odds scores of example QTL (BCa) for melanophore spacing (nearest neighbor distance, log2(DV:AP variation), and number of melanophore elements across 25 chromosomes. Variation segegrating between species in blue and within |
|
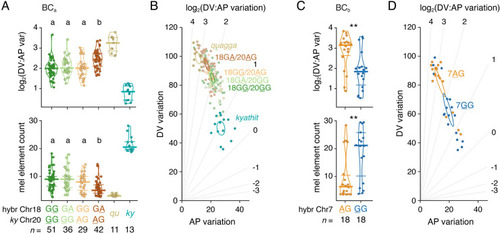
(A) Genotypes at Chr18:5246981 and Chr20:23025453 linked to variants affecting log2(DV:AP variation) and melanophore element count in BCa progeny. Genotypes are listed as maternally-inherited |