- Title
-
Dynamic optima in cell sizes during early development enable normal gastrulation in zebrafish embryos
- Authors
- Menon, T., Borbora, A.S., Kumar, R., Nair, S.
- Source
- Full text @ Dev. Biol.
|
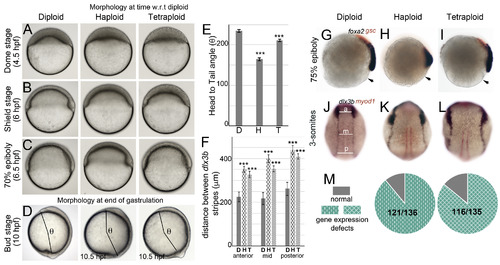
Embryos with average cell sizes different than the norm manifest epiboly delay and dorsal convergence defects during gastrulation. Live DIC images during epiboly and gastrulation (A–D). Haploids and tetraploids initiate epiboly (A) and gastrulation (B) later than diploids and develop with a delay during gastrulation (C). Haploids and tetraploids conclude gastrulation with a delay and manifest a shorter body axis at bud stage, quantified as the head to tail angle (beginning of head to tailbud from the center of the yolk ball, denoted as θ, D). Quantification of θ in diploids (n = 15), haploids (n = 10), tetraploids (n = 8) (E). RNA in situs for foxa2 (blue) and gsc (red) at 75% epiboly (G-I, diploid n = 34/34, haploid n = 62/63, tetraploid n = 58/66, arrow indicates embryo margin). RNA in situs for dlx3b (blue) and myod1 (red) at 3-somites (J-L, diploid n = 56/56, haploid n = 63/77, tetraploid n = 69/80). Quantification of the distance between dlx3b stripes along the AP axis at three sites anterior (a), mid (m) and posterior (p) as shown in J, for all conditions (F). Asterisks denote statistical significance based on unpaired t-test and One-way ANOVA (p ≤ 0.05), error bars represent Standard Error of Mean (SEM) for E and F. Cumulative quantification of all gastrulation defects (live and RNA in situ) in haploids and tetraploids (M). |
|
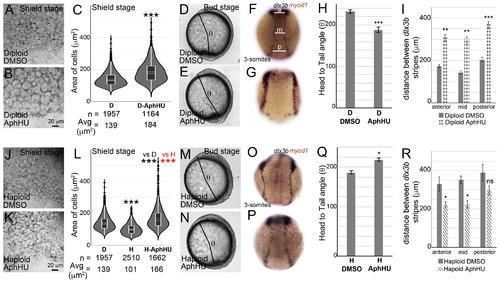
Gastrulation defects can be mimicked in diploids and rescued in haploids by increasing average cell areas.Diploids treated with DMSO (A, D, F) or AphHU (B, E, G). At shield stage, cell sizes in D-AphHU (B) are larger than in DMSO treated diploids (A). Cell area distributions, dotted horizontal line within box plots indicate mean value, n = number of correctly segmented cells that were used for area calculations from 3 embryos (C). D-AphHU embryos have a shorter body axis at bud stage, quantified as the head to tail angle (θ, D, E). Quantification of θ (H). RNA in situs for dlx3b (blue) and myod1 (red) at 3-somites for diploid DMSO (F, n = 29/29) and D-AphHU (G, n = 21/45). Quantification of the distance between dlx3b stripes along the AP axis at three sites anterior (a), mid (m) and posterior (p) as shown in F (I). Haploids treated with DMSO (J, M, O) or AphHU (K, N, P). At shield stages, cell sizes in H-AphHU (K) are larger than in DMSO treated haploids (J). Cell area distributions, dotted horizontal line within box plots indicate mean value, n = number of correctly segmented cells that were used for area calculations from 3 embryos (L). Body axis elongation quantified as θ, is rescued in H-AphHU (M, N, Q). RNA in situs for dlx3b (blue) and myod1 (red) at 3-somites for haploid DMSO (O, 16/26) and H-AphHU (P, n = 21/27). Quantification of the distance between dlx3b stripes along the AP axis at three sites anterior (a), mid (m) and posterior (p) as shown in F (R). Asterisks denote statistical significance based on Mann Whitney (C and L, P ≤ 0.001). Asterisks denote statistical significance based on unpaired t-test and One-way ANOVA (p ≤ 0.05), error bars represent Standard Error of Mean (SEM) for H, I, Q and R. |
|
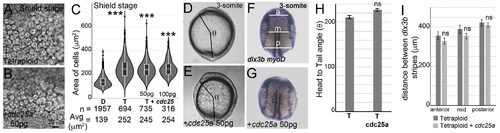
Overexpression of cdc25a does not reduce cell areas in tetraploidsUninjected tetraploids (A, D, F) and tetraploids injected with 50 pg of cdc25a mRNA (B, E, G). At shield stages, cell sizes in cdc25a injected tetraploid embryos (B) resemble uninjected tetraploid embryos (A). Cell area distributions, dotted horizontal line within box plots indicate mean value, n = number of correctly segmented cells that were used for area calculations from 3 embryos (C). Asterisks denote statistical significance based on Mann Whitney (p ≤ 0.05) compared to normal diploid embryos (C). Body axis elongation in cdc25a injected tetraploids resemble uninjected tetraploids at end of gastrulation (D, n = 28/28, E, n = 30/32 H), quantified as the head to tail angle (θ, D, E) quantification of θ (H). RNA in situs for dlx3b and myoD (blue) in uninjected tetraploids (F, n = 28/33) and cdc25a injected tetraploids (G, n = 24/29), show failure to converge dorsally. Quantification of the distance between dlx3b stripes along the AP axis at three sites anterior (a), mid (m) and posterior (p) as shown in F (I). Asterisks denote statistical significance based on unpaired t-test and One-way ANOVA (p ≤ 0.05), error bars represent Standard Error of Mean (SEM) for H and I. |
|
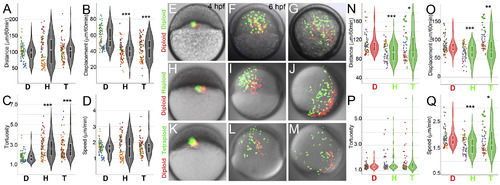
Cells from haploid and tetraploid embryos migrate suboptimally during gastrulation. Cell migration quantification from shield to bud stages of gastrulation in mosaic (A–D) and chimeric embryos (E–Q). For comparisons across experimental conditions and individual timelapses, the quantifications shown are for a normalized duration of 60 min. Graphs show the distribution of total distance travelled (A), displacement (B), tortuosity (C) and speed (D) of 90 cells from 3 diploid (D), 4 haploid (H) and 4 tetraploid (T) embryos. Diploid red cells co-transplanted with diploid green cells tend to mix during migration (E–G). Diploid (red) cells co-transplanted with haploid or tetraploid (green) cells into diploid hosts at sphere stage (4hpf, H, K) and imaged through gastrulation (I, J, L, M), in which the haploid and tetraploid cells are always farther away from the dorsal shield (D). Graphs show the distribution of total distance travelled (N), displacement (O) tortuosity (P) and speed (Q) of 80 diploid (D, red), haploid (H, green) and tetraploid (T, green) from 5 host embryos of each co-transplant. Asterisks denote statistical significance based on Mann Whitney (p < 0.0001, 0.001 and < 0.05) and dotted horizontal line within box plots indicate mean value. Dots represent individual cells and colours of the dots represent individual embryos quantified from independent experiments. |
Reprinted from Developmental Biology, 468(1-2), Menon, T., Borbora, A.S., Kumar, R., Nair, S., Dynamic optima in cell sizes during early development enable normal gastrulation in zebrafish embryos, 26-40, Copyright (2020) with permission from Elsevier. Full text @ Dev. Biol.