- Title
-
MultiFRAGing: Rapid and Simultaneous Genotyping of Multiple Alleles in a Single Reaction
- Authors
- Petree, C., Varshney, G.K.
- Source
- Full text @ Sci. Rep.
|
Overview of MultiFRAGing method. ( |
|
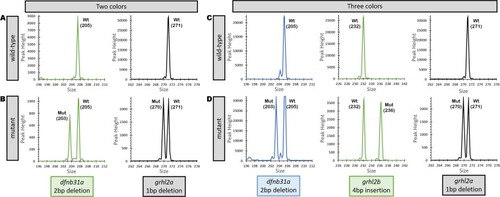
Fragment Analysis PCR plots from wild-type samples and a pool of samples amplified together. ( |
|
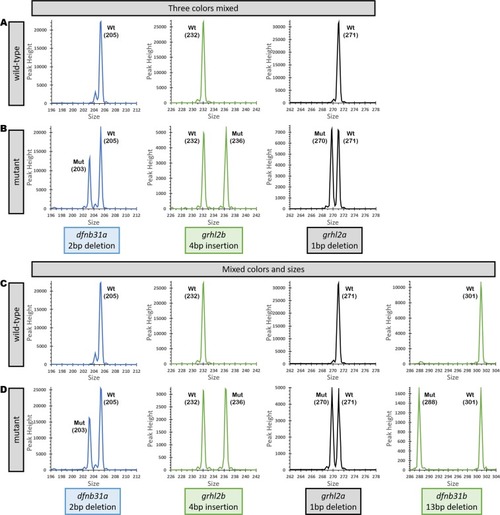
Fragment Analysis plots from wild-type controls and mutants from fragments separated by color, sizes and amplified individually, then pooled together for genetic analyzer. ( |
|
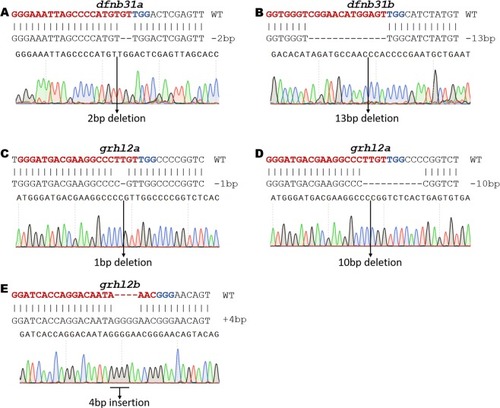
Validation of indels by Sanger Sequencing. Each indel that was identified by fragment analysis was sequenced using the Sanger method to establish the correlation between fragment analysis and Sanger sequencing Data. All indels from fragment analysis showed similar indel size in Sanger sequencing. ( |