- Title
-
Zebrafish modeling reveals that SPINT1 regulates the aggressiveness of skin cutaneous melanoma and its crosstalk with tumor immune microenvironment
- Authors
- Gómez-Abenza, E., Ibáñez-Molero, S., García-Moreno, D., Fuentes, I., Zon, L.I., Mione, M.C., Cayuela, M.L., Gabellini, C., Mulero, V.
- Source
- Full text @ J. Exp. Clin. Cancer Res.
|
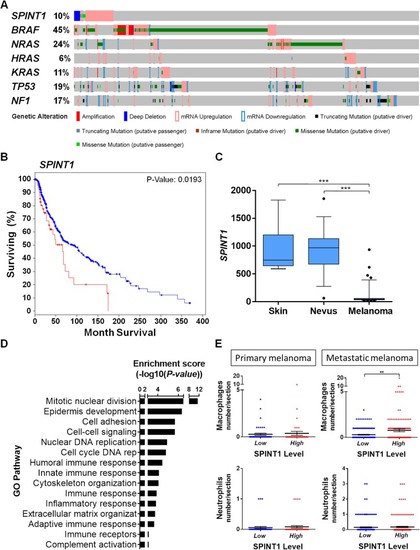
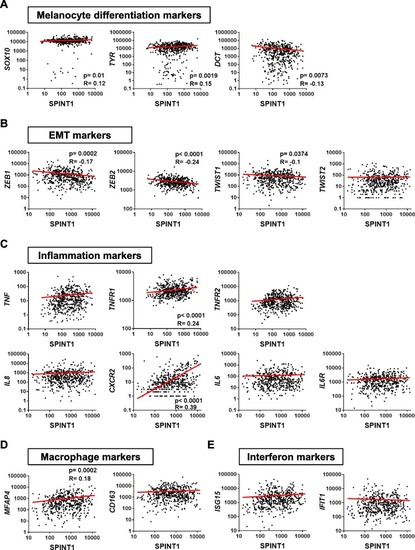
SPINT1 genetic alterations are associated with poor prognosis of SKCM patients. |
|
|
|
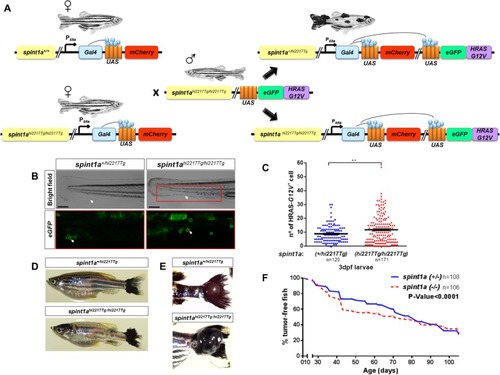
Inflammation accelerates the onset of SKCM in zebrafish. EXPRESSION / LABELING:
PHENOTYPE:
|
|
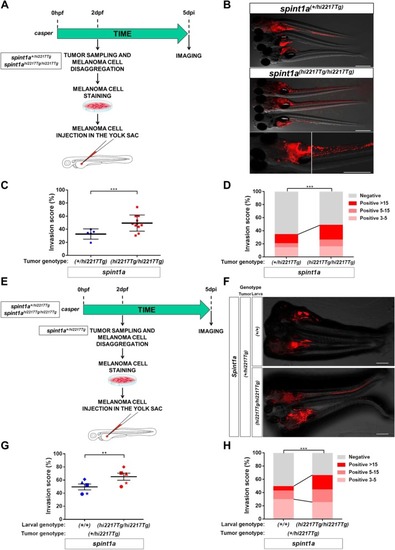
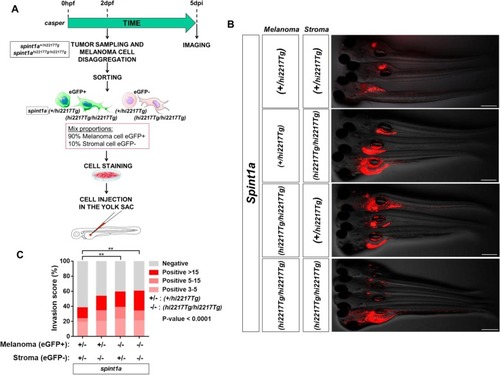
Spint1a deficiency is required at cell autonomous and non-autonomous levels to enhance SKCM cell dissemination in a zebrafish larval allotrasplantation model. Analysis of dissemination of control and Spint1a-deficient SKCM allotransplants in wild type larvae ( |
|
Spint1a deficiency both in stromal and tumor cell enhances SKCM dissemination in zebrafish larval model. |
|
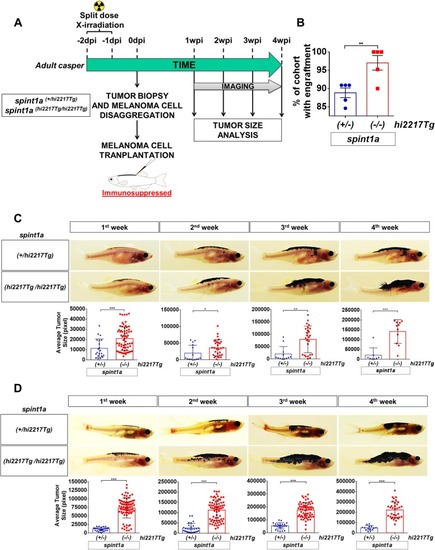
Spint1a-deficient SKCM shows enhanced aggressiveness in adult zebrafish allotransplantation assays. |