- Title
-
Biomechanical signaling within the developing zebrafish heart attunes endocardial growth to myocardial chamber dimensions
- Authors
- Bornhorst, D., Xia, P., Nakajima, H., Dingare, C., Herzog, W., Lecaudey, V., Mochizuki, N., Heisenberg, C.P., Yelon, D., Abdelilah-Seyfried, S.
- Source
- Full text @ Nat. Commun.
|
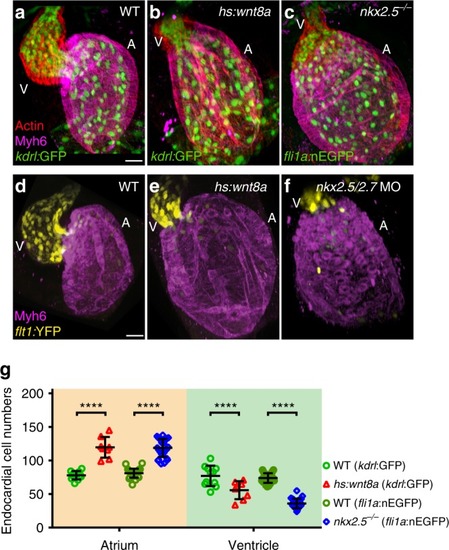
Overexpression of Wnt8a or loss of Nkx2.5 causes a shift in endocardial chamber proportions. |
|
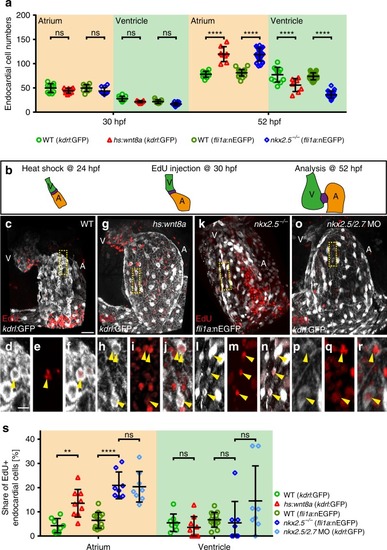
Atrial endocardial cell proliferation is increased upon Wnt8a overexpression or in |
|
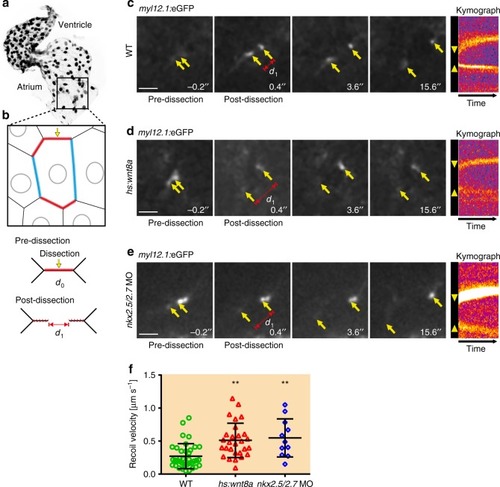
Endocardial tissue tension increases upon Wnt8a overexpression or loss of Nkx2.5/Nkx2.7. |
|
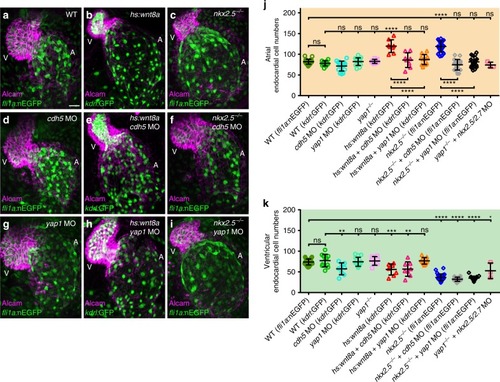
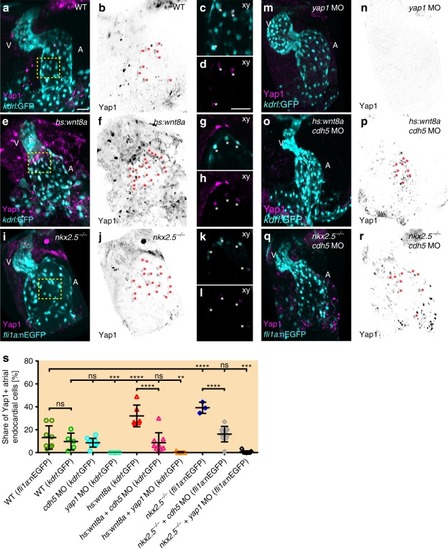
Loss of Cadherin-5 or Yap1 prevents endocardial cell number increases during atrial chamber expansion. |
|
Yap1 nuclear localization within the atrial endocardium increases upon chamber expansion. |