- Title
-
H3K27me3-mediated silencing of structural genes is required for zebrafish heart regeneration
- Authors
- Ben-Yair, R., Butty, V.L., Busby, M., Qiu, Y., Levine, S.S., Goren, A., Boyer, L.A., Burns, C.G., Burns, C.E.
- Source
- Full text @ Development
|
|
|
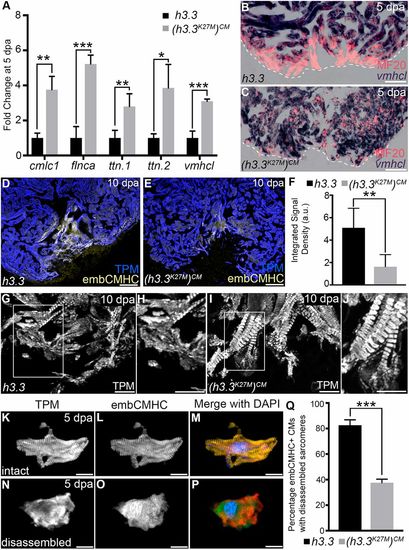
H3K27me3 deposition is required for sarcomere disassembly in wound edge cardiomyocytes. (A) Bar graph showing relative levels of sarcomeric transcripts in Tg(hsp70l:h3.3) and Tg(hsp70l:h3.3K27M)CM wound edge samples at 5 dpa, as measured by quantitative PCR (n=3 biological replicates for each cohort). Data are mean±s.d. *P<0.05; **P<0.01; ***P<0.001. (B,C) Section in situ hybridization for ventricular myosin (vmhcl) expression (blue) and antibody staining for myosin heavy chain (red). Control Tg(hsp70l:h3.3) hearts (B; n=6/6), but not Tg(hsp70l:h3.3K27M)CM hearts (C; n=5/5), exhibit reduced vmhcl expression in wound edge myocardium at 5 dpa. (D,E) Representative Tg(hsp70l:h3.3) (D) or Tg(hsp70l:h3.3K27M)CM (E) cardiac sections co-immunostained for tropomyosin (TPM; blue) and embCMHC (yellow). Tg(hsp70l:h3.3K27M)CM hearts showed relatively lower embCMHC signals at the wound edge compared with Tg(hsp70l:h3.3) hearts (n=8/8 and 11/11 hearts, respectively). (F) Bar graph showing the integrated signal densities of embCMHC in arbitrary units in Tg(hsp70l:h3.3) and Tg(hsp70l:h3.3K27M)CM hearts at 10 dpa. (G-J) Wound edge regions in cardiac sections immunostained for tropomyosin (TPM) at 10 dpa in Tg(hsp70l:h3.3) or Tg(hsp70l:h3.3K27M)CM hearts. Boxed regions in G and I are shown in H and J, respectively. Although wound edge cardiomyocytes show dissociated sarcomeres in Tg(hsp70l:h3.3) animals (n=5/5), sarcomere structure appears preserved in Tg(hsp70l:h3.3K27M)CM animals (n=4/6). (K-P) Split channel (K,L,N,O) or merged (M,P) confocal scans of dissociated cardiomyocytes immunostained for tropomyosin (TPM; red) and embryonic cardiac myosin heavy chain (embCMHC; green), and counterstained with DAPI (blue) to visualize sarcomere integrity in injury-responsive cardiomyocytes. (Q) The percentage of embCMHC+ cardiomyocytes with partially disassembled sarcomeres as shown in N-P in Tg(hsp70l:h3.3) (n=57 embCMHC+ CMs derived from nine apices) and Tg(hsp70l:h3.3K27M)CM (n=77 embCMHC+ CMs derived from nine apices) hearts. Data are mean±s.d. *P<0.05; **P<0.01; ***P<0.001. P-values were calculated using unpaired two-tailed Student's t-test. Scale bars: 100 µm in B-E; 10 µm in G-P. |
|
|