- Title
-
An SMN-Dependent U12 Splicing Event Essential for Motor Circuit Function
- Authors
- Lotti, F., Imlach, W.L., Saieva, L., Beck, E.S., Hao le, T., Li, D.K., Jiao, W., Mentis, G.Z., Beattie, C.E., McCabe, B.D., and Pellizzoni, L.
- Source
- Full text @ Cell
|
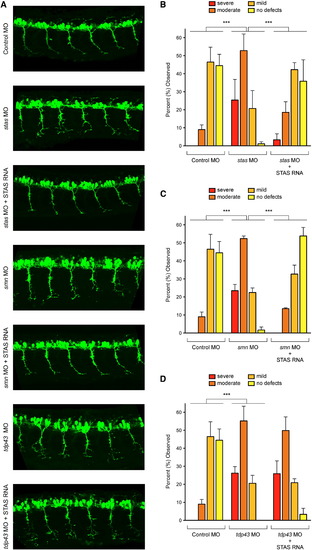
Stasimon Is Required for Motor Neuron Development and Rescues SMN-Dependent Motor Axon Defects in Zebrafish Embryos(A) Representative lateral views of motor axons in Tg(mnx1:GFP) zebrafish embryos expressing GFP in motor neurons and injected with control MO, as well as stas MO, smn MO, and tdp43 MO either with or without coinjected human STAS RNA.(B) Quantification of the effects of Stasimon deficiency on motor axon development in zebrafish. Motor axons were scored in Tg(mnx1:GFP) embryos injected with control MO, stas MO, or stas MO + STAS RNA. Embryos were classified as severe, moderate, mild, or no defects based on the severity of motor axon defects, and the percentage for each group is shown.(C) Quantification of Stasimon effects on SMN-dependent motor axon defects in zebrafish. Motor axons were scored in Tg(mnx1:GFP) embryos injected with control MO, smn MO, or smn MO + STAS RNA and embryos were classified as in (B).(D) Quantification of Stasimon effects on TDP43-dependent motor axon defects in zebrafish. Motor axons were scored in Tg(mnx1:GFP) embryos injected with control MO, tdp43 MO, or tdp43 MO + STAS RNA, and embryos were classified as in (B).Data in all graphs are represented as mean and SEM. See also Figure S6. |
|
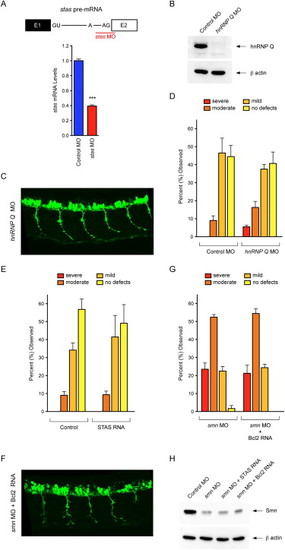
Specificity of Stasimon Effects on Motor Axon Development in Zebrafish Embryos, Related to Figure 6(A) Schematic representation of a portion of the zebrafish stasimon pre-mRNA that includes the splice junction between intron 1 and exon 2 targeted by stas MO. Splice sites (GU and AG) and branch point adenosine (A) are indicated (top panel). RT-qPCR analysis of Stasimon mRNA levels in zebrafish Tg(mnx1:GFP) embryos injected with Control MO and stas MO normalized to β-actin mRNA (bottom panel). Data are represented as mean and SEM.(B) Western blot analysis of hnRNP Q protein levels in Tg(mnx1:GFP) embryos injected with Control MO and hnrnp Q MO.(C) Representative lateral view of motor axons in zebrafish Tg(mnx1:GFP) morphants injected with hnrnp Q MO.(D) Quantification of the effects of hnRNP Q deficiency on motor axon development in zebrafish. Motor axons were scored in Tg(mnx1:GFP) embryos injected with Control MO or hnrnp Q MO. Embryos were classified as severe, moderate, mild or no defects based on the severity of motor axon defects, as previously described (Carrel et al., 2006), and the percentage for each group is shown. Data are represented as mean and SEM.(E) Quantification of the effects of Stasimon overexpression on normal motor axon development in zebrafish. Motor axons were scored in Tg(mnx1:GFP) embryos injected with either control MO or STAS RNA and embryos were classified as in (D). Data are represented as mean and SEM.(F) Representative lateral view of motor axons in zebrafish Tg(mnx1:GFP) morphants co-injected with smn MO and Bcl2 RNA.(G) Quantification of Bcl2 effects on SMN-dependent motor axon defects in zebrafish. Motor axons were scored in Tg(mnx1:GFP) embryos injected with smn MO or smn MO together with Bcl2 RNA and embryos were classified as in (D). Data are represented as mean and SEM.(H) Western blot analysis of Smn protein levels in Tg(mnx1:GFP) embryos injected with Control MO as well as smn MO either in the presence or in the absence of co-injected STAS RNA and Bcl2 RNA. |
Reprinted from Cell, 151(2), Lotti, F., Imlach, W.L., Saieva, L., Beck, E.S., Hao le, T., Li, D.K., Jiao, W., Mentis, G.Z., Beattie, C.E., McCabe, B.D., and Pellizzoni, L., An SMN-Dependent U12 Splicing Event Essential for Motor Circuit Function, 440-454, Copyright (2012) with permission from Elsevier. Full text @ Cell