PUBLICATION
Optimal tagging strategies for illuminating expression profiles of genes with different abundance in zebrafish
- Authors
- Liu, J., Li, W., Jin, X., Lin, F., Han, J., Zhang, Y.
- ID
- ZDB-PUB-231222-10
- Date
- 2023
- Source
- Communications biology 6: 13001300 (Journal)
- Registered Authors
- Keywords
- none
- MeSH Terms
-
- Fluorescence
- Genome
- Animals
- CRISPR-Cas Systems*
- Zebrafish*/genetics
- PubMed
- 38129658 Full text @ Commun Biol
Abstract
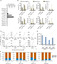
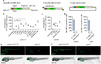
CRISPR-mediated knock-in (KI) technology opens a new era of fluorescent-protein labeling in zebrafish, a preferred model organism for in vivo imaging. We described here an optimized zebrafish gene-tagging strategy, which enables easy and high-efficiency KI, ensures high odds of obtaining seamless KI germlines and is suitable for wide applications. Plasmid donors for 3'-labeling were optimized by shortening the microhomologous arms and by reducing the number and reversing the sequence of the consensus Cas9/sgRNA binding sites. To allow for scar-less KI across the genome, linearized dsDNA donors with 5'-chemical modifications were generated and successfully incorporated into our method. To refine the germline screen workflow and expedite the screen process, we combined fluorescence enrichment and caudal-fin junction-PCR. Furthermore, to trace proteins expressed at a low abundance, we developed a fluorescent signal amplifier using the transcriptional activation strategy. Together, our strategies enable efficient gene-tagging and sensitive expression detection for almost every gene in zebrafish.
Genes / Markers
Expression
Phenotype
Mutations / Transgenics
Human Disease / Model
Sequence Targeting Reagents
Fish
Orthology
Engineered Foreign Genes
Mapping