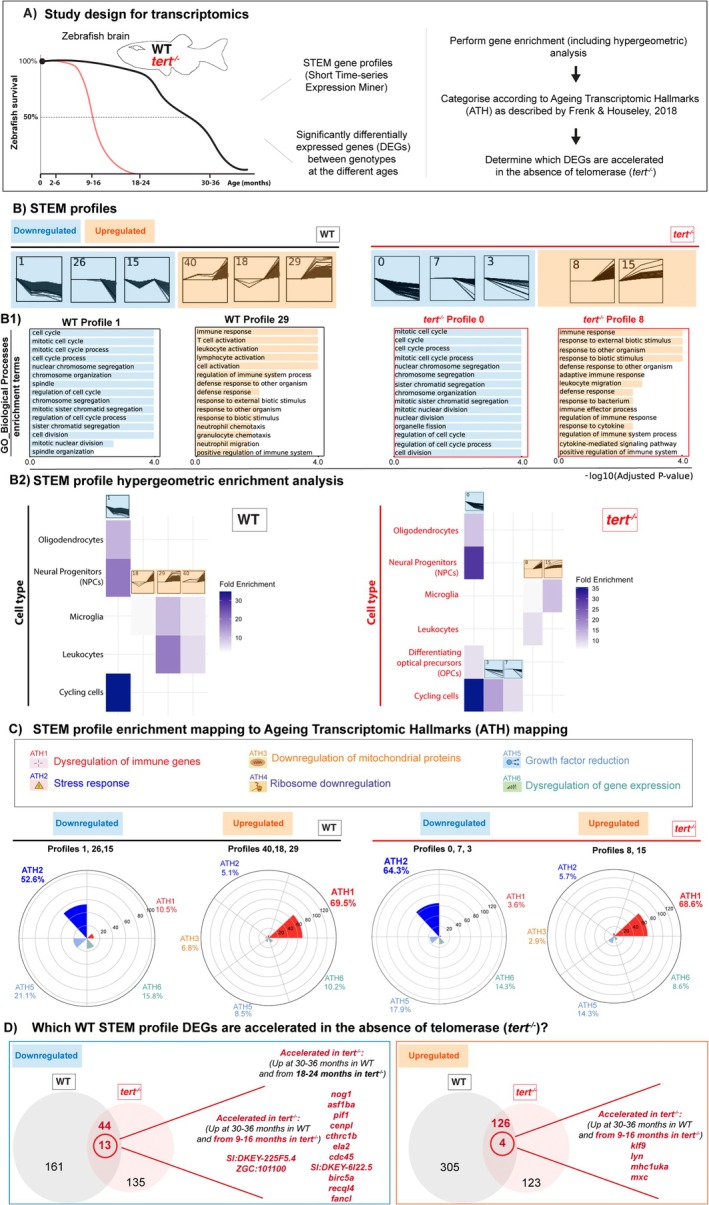
FIGURE 1
Transcriptomic signatures of ageing in the presence and absence of telomerase in the zebrafish brain. (A) Schematic figure of the study design for transcriptomics: RNA from whole brain tissue from young (2–6 months), young adult (9–16 months), middle aged (18–24 months) and old, (30–36 months) WT fish, and young (2–6 months), middle aged (9–16 months) and old (18–24 months)