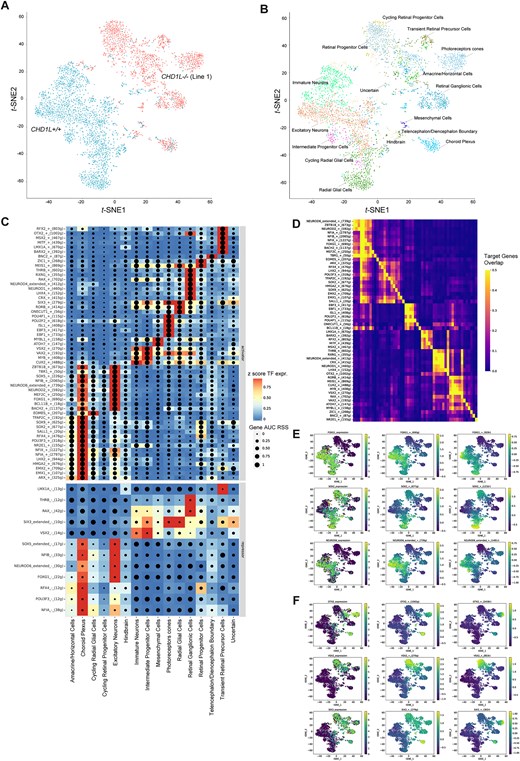
Fig. 8 SCENIC + analysis reveals CHD1L function in forebrain cell-fate determination. (A) t-SNE dimensionality reduction of 4019 cells based on gene and target region enrichment scores of eRegulons. Cells are colored according to their genotypes CHD1L+/+ and CHD1L−/− and were analyzed for gene expression and chromatin accessibility in 60-day in vitro hCO. (B) t-SNE dimensionality reduction of organoid cells based on gene and target region enrichment scores of eRegulons. Cells are colored according to their identity. (C) Heatmap/dot-plot showing transcription factor (TF) expression of the eRegulon on a color scale and cell-type specificity (RSS) of the eRegulon on a size scale. Cell types are ordered on the basis of their gene expression similarity. (D) Overlap of target genes of eRegulons. The overlap is divided by the number of target genes of the eRegulon in each row. (E) Representative t-SNE dimensionality reduction of multiome dataset colored based on TF expression, target gene and region activity of eRegulons for telencephalic markers (FOXG1, SOX2, NEUROD6). (F) Representative t-SNE dimensionality reduction of multiome dataset colored based on TF expression, target gene and region activity of eRegulons for retinal markers (OTX2, VSX2, SIX3). AUC, area under the recovery curve; g, gene; r, region; RSS, eRegulon specificity score; TF, transcription factor.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Nucleic Acids Res.