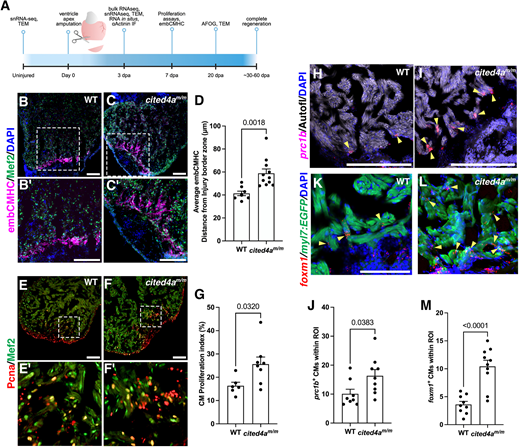
Fig. 4 Cited4a limits CM dedifferentiation and proliferation in heart regeneration. (A) Timeline of injury and experiments. dpa, days post amputation. Created in BioRender by Forman-Rubinksy, R. (2025) https://BioRender.com/ifbq3md. This figure was sublicensed under CC-BY 4.0 terms. (B-C′) Immunostaining of embCMHC (magenta) to mark dedifferentiating CMs and Mef2a/c (green) to mark CMs, with DAPI (blue), in 3 dpa WT (B,B′) and cited4am/m (C,C′) ventricles. (D) Graph showing average distance of embCMHC staining from injury border zone in WT and cited4am/m hearts. WT n=8; cited4am/m n=11. (E-F′) Immunostaining of Pcna (red) to mark proliferating CMs and Mef2 (green) to mark CMs in 3 dpa WT (E,E′) and cited4am/m (F,F′) ventricles. (G) Quantification of CM proliferation index (Pcna+;Mef2+/Total Mef2c+). WT n=6; cited4am/m n=8. (H,I) prc1b (red) transcripts detected at the injury border zone by RNAscope with DAPI and autofluorescent myocardial tissue in 3 dpa WT (H) and cited4am/m (I) ventricles. (J) Quantification of proliferating prc1b+ nuclei in the myocardium. WT n=8; cited4am/m n=9. (K,L) foxm1 (red) transcripts detected at the injury border zone by RNAscope with myl7:EGFP to mark CMs, and DAPI, in 3 dpa WT (K) and cited4am/m (L) ventricles. Arrowheads highlight examples of proliferating CM in image. (M) Quantification of foxm1+ CMs at the injury border zone. WT n=9; cited4am/m n=10. Data are mean±s.e.m. Unpaired two-tailed t-test. Scale bars: 100 µm.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development