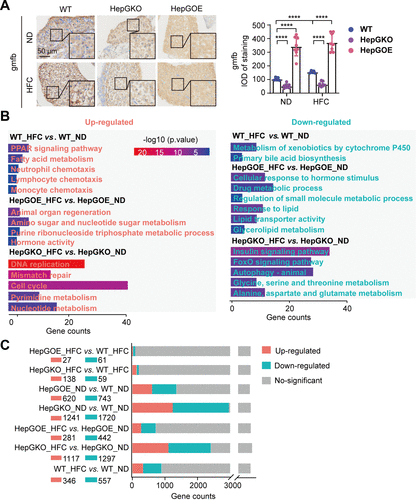
Fig. 4 Transcriptome analysis of zebrafish in MASLD liver. A: representative anti-gmfb immunochemical images in liver slices from ND and HFC groups of WT, HepGKO, and HepGOE zebrafish, and quantification of the gmfb-positive staining density. Scale bar = 50 μm. n = 8 fish per group. Experiments were repeated on at least 3 clutches. One-way ANOVA, ****P < 0.0001 vs. ND or WT group. B: GO and KEGG analyses of transcriptome data associated with comparison of WT_HFC vs. WT_ND, HepGOE_HFC vs. HepGOE_ND, and HepGKO_HFC vs. HepGKO_ND. n = 2 samples for each group, 10 fish per sample. Significant functional entries were identified with criteria of P value < 0.05. C: bar plot shows the differential gene expression of transcriptome data defined with criteria of |log2(Fold Change)| > 1 and Padj < 0.05. n = 2 samples for each group, 10 fish per sample. gmfb, glia maturation factor-β; GO, Gene Ontology; hep-gmfb, gmfb in hepatocyte; HepGKO, hepatocyte-specific gmfb knockout; HepGOE, hepatocyte-specific gmfb overexpression; HFC, high fat, high cholesterol diet; IOD, integral optical density; KEGG; Kyoto Encyclopedia of Genes and Genomes; MASLD, metabolism-associated steatotic liver disease; ND, normal diet; WT, wild-type.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Am. J. Physiol. Gastrointest. Liver Physiol.