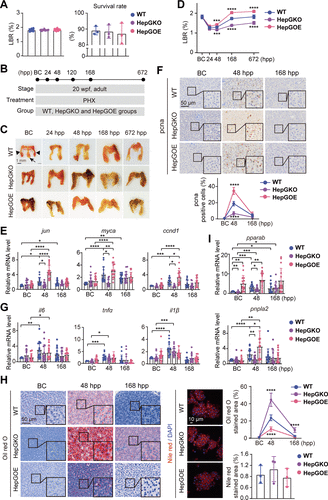
Fig. 2 Hep-gmfb promotes liver regeneration and prevents hepatic steatosis post PHX. A: LBR and survival rate of zebrafish from WT, HepGKO, and HepGOE groups. For the LBR, n = 8 samples, one fish at 20 wpf per sample. For the survival rate analysis, n = 3 replicated groups, 100 fish at 5 dpf per group. B: schematic illustration of the construction of zebrafish PHX model with WT, HepGKO, and HepGOE lines. C: representative images of livers from WT, HepGKO, and HepGOE groups at indicated timepoints post-PHx. In BC panel, the black arrowheads indicate the two lateral lobes, the black arrow indicates the ventral lobe, and the red short, dashed line indicates the PHX incision site. Scale bar = 1 mm. n = 8 fish per group. D: LBR of zebrafish from WT, HepGKO, and HepGOE groups at indicated timepoints post-PHx. n = 8 fish per group. One-way ANOVA, ***P < 0.001, ****P < 0.0001 vs. WT group. E, G, and I: expression levels of indicated genes jun, myca, and ccnd1 in (E); il6, tnfα, and il1β in (G); and pparab and pnpla2 in (I) in liver samples from WT, HepGKO, and HepGOE groups at BC, 48 hpp and 168 hpp. n = 15 fish per group. Data were presented as relative expression levels. One-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 vs. BC or WT group. F: representative anti-pcna immunochemical images in liver sections and quantification of pcna positive staining cell from WT, HepGKO, and HepGOE groups at BC, 48 hpp and 168 hpp. Scale bar = 50 μm. n = 8 fish per group. One-way ANOVA, ****P < 0.0001 vs. WT group. H: representative images of lipid droplets with oil red O staining in liver slices of PHX model, and the Nile red staining lipid droplets in zebrafish primary hepatocytes from WT, HepGKO, and HepGOE groups, and quantification of the positive staining areas. Scale bar = 50 μm for oil red O staining and 10 μm for Nile red staining. n = 12 samples for oil red O staining, 1 fish per sample. n = 8 fish per group for Nile red staining. One-way ANOVA, ****P < 0.0001 vs. WT group. All experiments were repeated on at least 3 clutches. BC, baseline control; ccnd1, cyclin D1; DAPI, 4′,6-diamidino-2-phenylindole; gapdh, glyceraldehyde-3-phosphate dehydrogenase; hep-gmfb, gmfb in hepatocyte; HepGKO, hepatocyte-specific gmfb knockout; HepGOE, hepatocyte-specific gmfb overexpression; hpp, hours post-PHX; il1β, interleukin 1 beta; il6, interleukin 6; jun, Jun proto-oncogene, AP-1 transcription factor subunit; LBR, liver-to-body weight ratio; myca, MYC proto-oncogene, bHLH transcription factor a; pcna, proliferating cell nuclear antigen; PHX, partial hepatectomy; pparab, peroxisome proliferator-activated receptor alpha b; pnpla2, patatin-like phospholipase domain containing 2; tnfα, tumor necrosis factor alpha; wpf, weeks post fertilization; WT, wild-type.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Am. J. Physiol. Gastrointest. Liver Physiol.