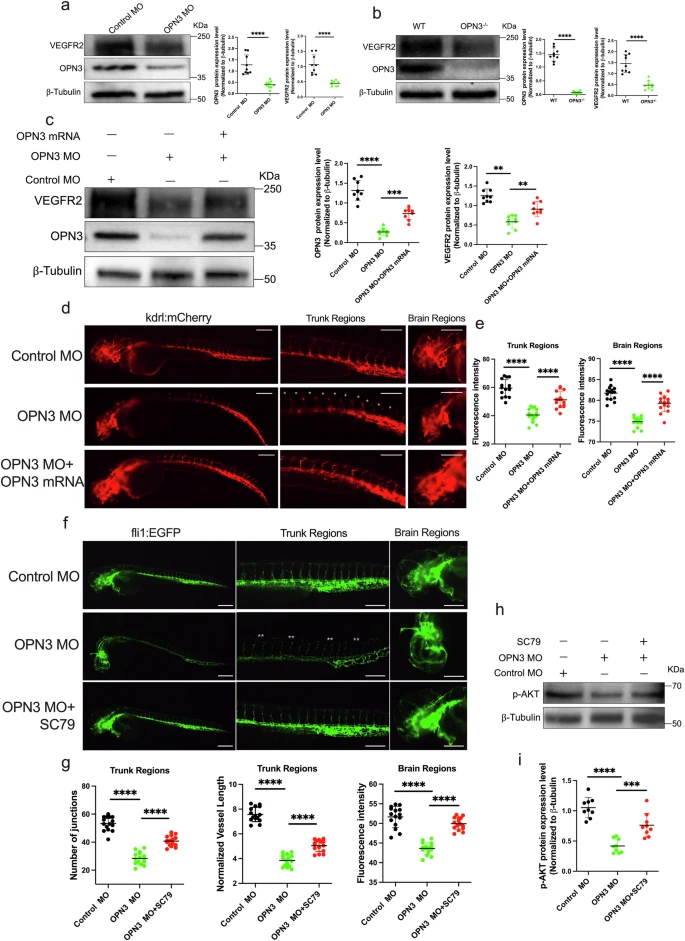
Fig. 8 OPN3 regulates VEGFR2 expression and AKT activation in vivo.a Western blot analysis of OPN3 and VEGFR2 protein expression in fli1:EGFP zebrafish embryos after OPN3 knockdown using MO. β-tubulin was used as a loading control for normalization in the WB analysis. Relative protein levels were quantified using ImageJ (n = 9 batches of injected zebrafish embryos, each batch containing 30 embryos). Statistical analysis was performed using an unpaired t-test: ****p < 0.0001. b Western blot analysis of OPN3 and VEGFR2 protein expression in WT and OPN3-/- at 48 hpf. β-tubulin was used as a loading control for normalization in the WB analysis. Relative protein levels were quantified using ImageJ (n = 9 batches of injected zebrafish embryos, each batch containing 30 embryos). Statistical analysis was performed using an unpaired t-test: ****p < 0.0001. c Western blot analysis of OPN3 and VEGFR2 protein expression in kdrl:mCherry zebrafish embryos after OPN3 knockdown using MO, and after co-injection of OPN3 MO with OPN3 mRNA. β-tubulin was used as a loading control for normalization in the WB analysis. Relative protein levels were quantified using ImageJ (n = 9 batches of injected zebrafish embryos, each batch containing 30 embryos). Statistical analysis was performed using an unpaired t-test: **p < 0.01, ***p < 0.001, ****p < 0.0001. d Inject kdrl:mCherry transgenic zebrafish embryos with Control MO, OPN3 MO, or OPN3 MO and OPN3 mRNA, and image the treated embryos from each group at 48 hpf using a confocal microscope. The fluorescence of kdrl is absent and discontinuous in many places (*). e Use ImageJ to analyze the fluorescence intensity in the images from each group (n = 15, each individual data point represents the trunk or brain region of a single zebrafish embryo). Statistical analysis was performed using an unpaired t-test: ****p < 0.0001. The scale bar represents 100 μm. f Inject fli1:EGFP transgenic zebrafish embryos with Control MO, OPN3 MO, or OPN3 MO and SC79, and image the treated embryos from each group at 48 hpf using a confocal microscope. DLAVs and ISVs were absent and discontinuous in many places (**). g Use ImageJ to analyze the normalized vessel length and the number of junctions in the images from each group (n = 15, each individual data point represents the trunk or brain region of a single zebrafish embryo). Statistical analysis was performed using an unpaired t-test: ****p < 0.0001. The scale bar represents 100 μm. h Western blot analysis of p-AKT protein expression in fli1:EGFP zebrafish embryos after OPN3 knockdown using MO, and after co-injection of OPN3 MO with SC79. β-tubulin was used as a loading control for normalization in the WB analysis. i Relative protein levels were quantified using ImageJ (n = 9 batches of injected zebrafish embryos, each batch containing 30 embryos). Statistical analysis was performed using an unpaired t-test: ***p < 0.001, ****p < 0.0001. Data are presented as mean ± SEM.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Commun Biol