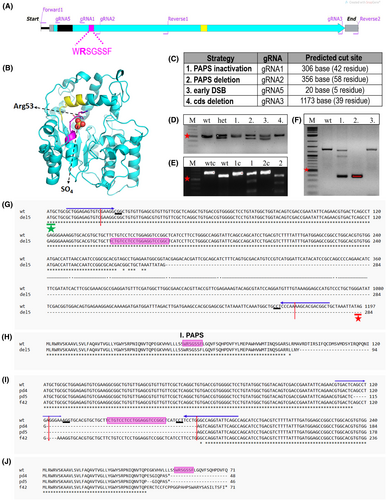
Fig. 2 chst6 mutagenesis strategy. (A) Graphical representation of chst6 gene cds. Carbohydrate sulfotransferase domain (turquoise), 1st PAPS (magenta), and 2nd PAPS (yellow) and transmembrane (black) domains are colored. Primers used for generating the PCR product for genotyping of PAPS domain mutants (forward 1, reverse 1); cds deletion mutant or Sanger sequencing (forward 1, reverse 2); and gRNA binding sites are indicated. (B) Model of zebrafish CHST6 sulfotransferase domain indicating localization of 1st PAPS (a.a. 49–55, magenta) and 2nd PAPS (a.a. 202–210, yellow) and SO4 interaction with Arg53. Model was generated with the pymol Software. (C) Mutagenesis strategies and used gRNA annotations are shown. gRNA1 was used for the inactivation of the 1st PAPS domain by indel generation, gRNA1 and gRNA2 were used together to induce deletion of the 1st PAPS domain, gRNA5 was used to induce early indel before the transmembrane domain, and gRNA3 and gRNA5 were used together to induce deletion of the coding sequence. (D–F) Representative agarose gel images of genotyping analyses; (D) PAPS deletion (pd) mutation leads to generation of a 50 bp shorter mutant fragment. Lanes: marker, wt, heterozygous mutant, test fish 1, 2, 3, and 4 PCR products, (E) gRNA1 induced indel/frameshift (f) mutations were detected with T7EI assay. Presence of mutant DNA strand leads to formation of a mismatch and digestion by T7E1. Lanes: marker, wt control, wt digested, fish 1 control, fish 1 digested, fish 2 control, fish 2 digested PCR products. (F) F1 embryos obtained from founder fish of the cds deletion (del5) mutation were genotyped with PCR. chst6 genomic region was amplified with forward 1 and reverse 2 primers. Heterozygous embryo 1 had 3 bands (wt, mutant, and heteroduplex), whereas embryo 2 had only the short band, and nonmutant embryo 3 had the wt DNA band. Short band was expected to be 150–160 bp, but a longer band was detected. Red asterisk shows 500 bp marker band in all gels. (G–J) Sequence alignments of obtained mutants with wt chst6 (NM_001130621) were generated with clustal omega web tool. gRNA binding sequences are indicated with arrows on top, predicted DSB site is indicated with red vertical arrow, 1st PAPS coding sequence is highlighted with magenta, PAM sequence is underlined, and start codon is indicated with a green asterisk. (G) DNA sequence, (H) protein sequence alignment of del5 mutant to wt, multiple sequence alignment of (I) DNA sequence, (J) protein sequence of homozygous mutants (chst6pd4/pd4, chst6pd5/pd5, and chst6f42/f42) with wt.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ FEBS J.