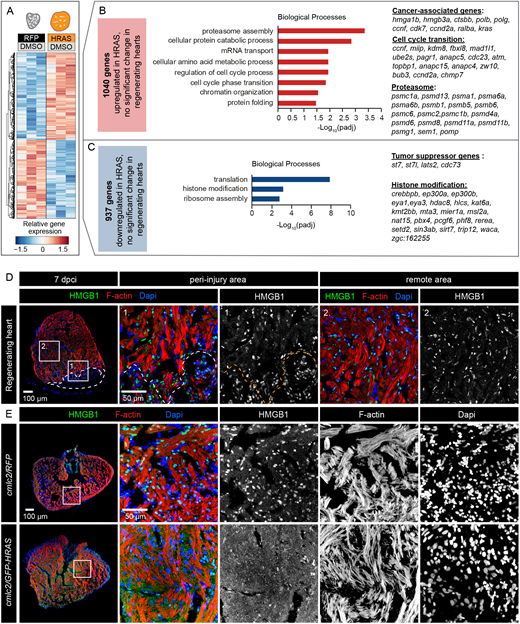
Fig. 6 Unique DE genes of HRAS-OE hearts compared with regenerating hearts. (A) Heatmap of log2-transformed normalized expression values for genes exclusively deregulated in cmlc2/GFP-HRAS compared with cmlc2/RFP, without significant changes in 7 dpci compared with uninjured hearts. (B) Selected enriched gene ontology terms for the 1040 genes exclusively upregulated in HRAS-induced hearts. Cancer-associated, cell cycle transition and proteasome genes are indicated. (C) Selected enriched gene ontology terms for the 937 genes exclusively downregulated in HRAS-induced hearts. Examples of tumor suppressor genes and histone modifiers are listed. (D,E) Immunostaining of regenerating and HRAS-expressing hearts reveals differential subcellular localization of HMGB1. Only in HRAS-OE hearts is this protein non-nuclear. n=4.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development