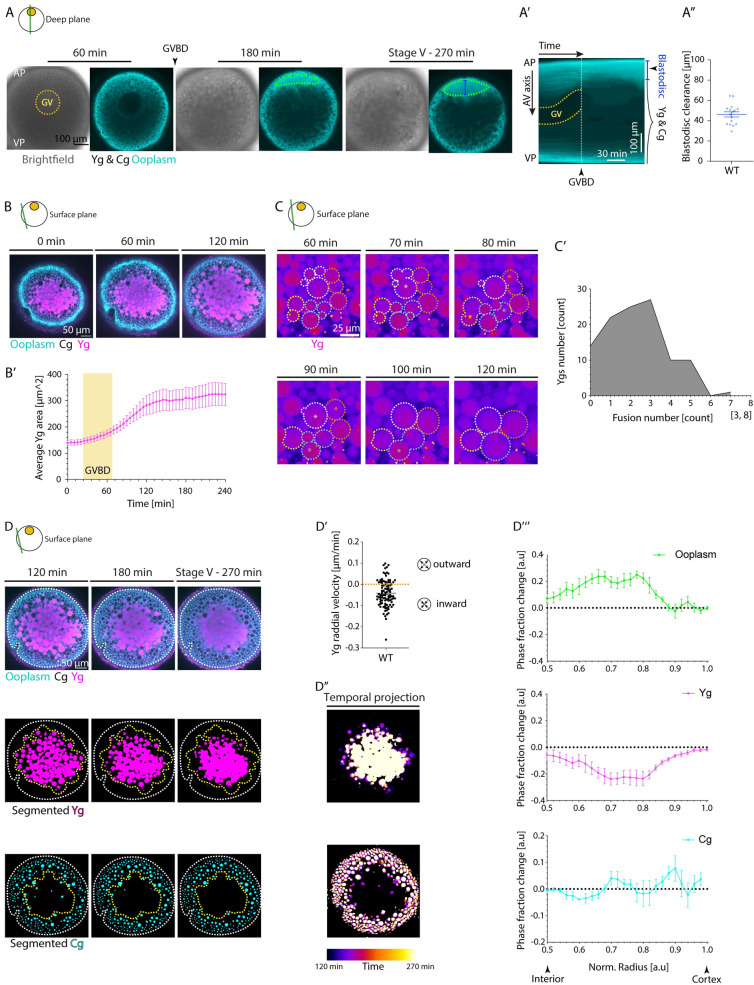
Fig 1
(A) Brightfield (left) and fluorescence (right) images of stage III Tg(hsp:clip170-GFP) oocytes labeling the ooplasm during oocyte maturation at 60, 180, and 270 min after maturation induction with the DHP hormone. Dashed yellow circle highlights the GV contour, and arrowhead denotes the GVBD onset. The green ROIs indicate the blastodisc region, and blue line marks the blastodisc height as measured in Fig 1A”. Ygs and Cgs are depicted by their exclusion of ooplasmic signal. (