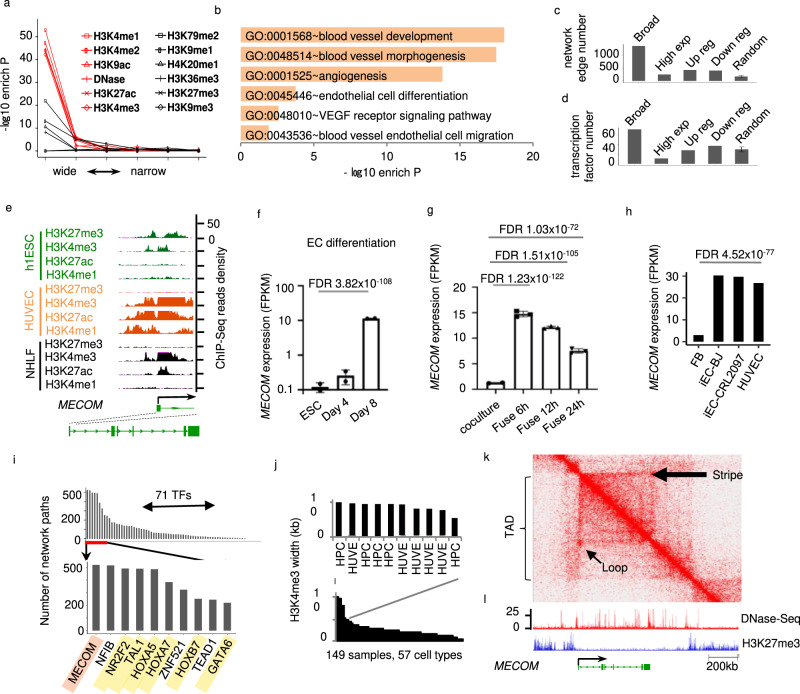
Fig. 1
a Enrichment of EC identity regulators plotted against the width of enrichment peak for each chromatin mark. b Pathways enriched in EC putative identity regulators defined by broad enrichment of activating histone modifications. c Number of regulation network edges connected to and (d) Number of transcription factors in EC putative identity regulators or other gene groups, of which each has the same number of genes. e Signal density of individual chromatin marks at the MECOM locus in h1ESC, HUVEC and NHLF. f–h Expression level of MECOM at individual intervals during EC differentiation mESC (f), the formation of mESC-hEC heterokayon (g), and human fibroblast to EC transdifferentiation (h). i Number of gene regulation network paths connected by each transcription factor (TFs). MECOM is highlighted in red and known EC regulators are highlighted in yellow. Gene regulation network for EC was defined by algorithm and database of the software CellNet. j H3K4me3 peak width at MECOM locus in individual ChIP-Seq samples. Sample information is provided in Supplementary Data 4. k Two-dimensional heat map showing the chromosomal contact frequency in HUVEC around MECOM locus. l Signal density of DNase-Seq and H3K27me3 ChIP-Seq in HUVEC around MECOM locus. Broad: broad active histone modifications; High exp: high expression; Up reg or Down reg: up or downregulated in EC relative to ESC. Random: randomly selected genes. Data are presented as mean values ± SD (f, g). n = 2 biologically independent samples (f), n = 3 biologically independent samples (g). P values are determined by two-tailed Fisher Exact test implemented in R v4.0.2 (a), modified two-tailed Fisher Exact test implemented in DAVID v6.8 (b), two-tailed Negative Binomial test (f, g), and two-tailed Poisson test implemented in edgeR v3.14.0 (h). EC data were from HUVECs (See Supplementary Data 4).