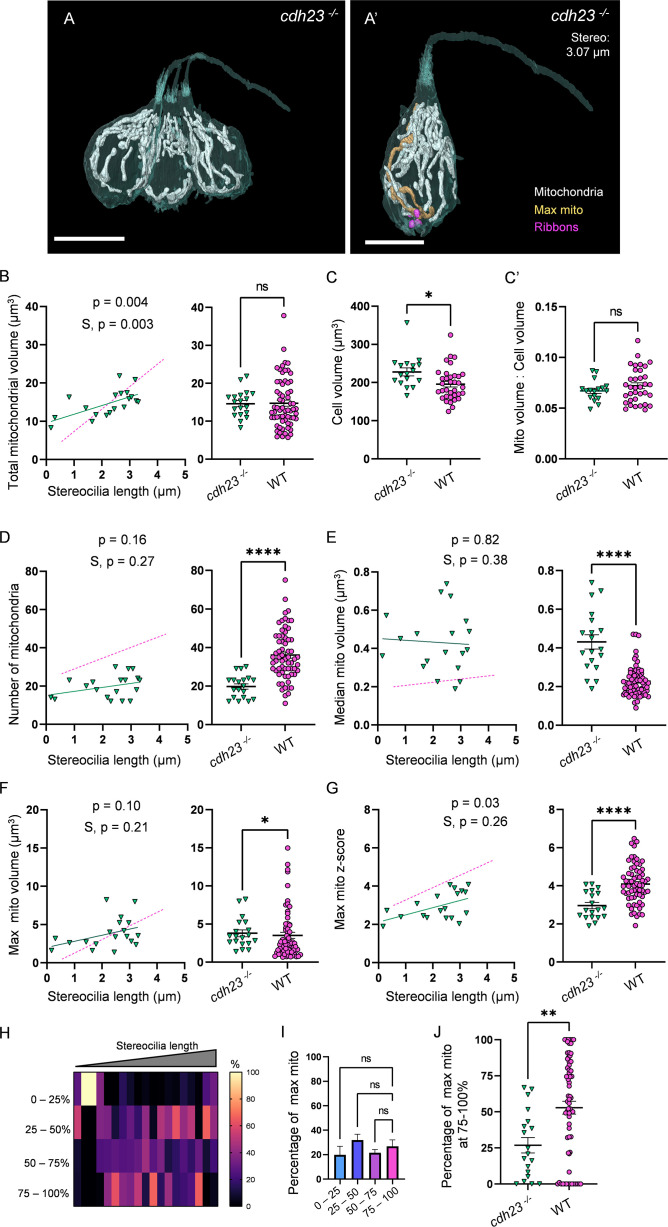
Figure 6
(A) Three representative cdh23 hair cells (HCs) with mitochondria labeled in white (from NM11, Figure 6—source data 2). Scale bar = 9.5 µm. (A’) Representative mature cdh23 HC (NM11), max mito shown in gold, synaptic ribbons shown in purple. Scale bar = 6 µm. (B) Comparison of total mitochondrial volume development (left) and average (right) between cdh23 and WT HCs. On average (in µm3): cdh23:14.6 ± 0.8; WT: 14.8 ± 0.8. Kolmogorov–Smirnov test, p=0.35 (C) Total HC volume for cdh23 and WT HCs. (In µm3) cdh23: 228.0 ± 10.3; WT: 195.6 ± 7.2. Kolmogorov–Smirnov test, p=0.03. (C’) Ratio of total mitochondrial volume to HC volume. cdh23: 0.07 ± 0.002; WT: 0.07 ± 0.003. Kolmogorov–Smirnov test, p=0.23. (D) Comparison of the number of HC mitochondria over development (left) and on average (right) in cdh23 and WT HCs. On average: cdh23: 19.8 ± 1.4; WT: 36.1 ± 1.6. Kolmogorov–Smirnov test, p<0.0001. (E) Comparison of the median mitochondrial volume over development (left) and on average (right). On average (in µm3): cdh23: 0.4 ± 0.04; WT: 0.2 ± 0.01. Kolmogorov–Smirnov test, p<0.0001. (F) Comparison of the max mito volume over development (left) and on average (right). On average (in µm3): cdh23: 3.8 ± 0.5; WT: 3.5 ± 0.4. Kolmogorov–Smirnov test, p=0.04. (G) Comparison of the max mito z-score in cdh23 and WT HCs over development (left) and on average (right). On average: cdh23: 3.0 ± 0.2; WT: 4.1 ± 0.1. Standard unpaired t-test, p<0.0001. (B–G) Solid line represents the standard linear regression for cdh23 HCs. Dashed line represents standard regression for WT HCs as in Figure 2. Significance of the cdh23 regression and differences in the slope from the WT regression (S) are indicated. (H) The percentage of the max mito located within each quadrant of cdh23 HCs represented as a heat map. (I) Summary of the heat map data shown in (H). Most apical quadrant (0–25%): 19.8 ± 7.0%; 25–50%: 31.9 ± 4.8%; 50–75%: 21.6 ± 2.6%; Most basal quadrant (75–100%): 26.8 ± 5.3%. Kruskal–Wallis test with Dunn’s multiple comparisons, nonsignificant. (J) Percentage of max mito located within the most basal quadrant for individual HCs. cdh23: 26.8 ± 5.3%, WT: 52.8 ± 4.4%. Kolmogorov–Smirnov test, p=0.008. (B, D–J) cdh23 data: n = 19 HCs, 4 NMs, 2 fish. WT data: n = 65 HCs, 5 NMs, 3 fish. (C, C’) cdh23 data: n = 17 HCs, 4 NMs, 2 fish. WT data: n = 35 HCs, 3 NMs, 3 fish. Where applicable, data are presented as the mean ± SEM.
Mechanotransduction is not necessary for high mitochondrial volume, but is required for development of mitochondrial architecture.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife