Figure 5
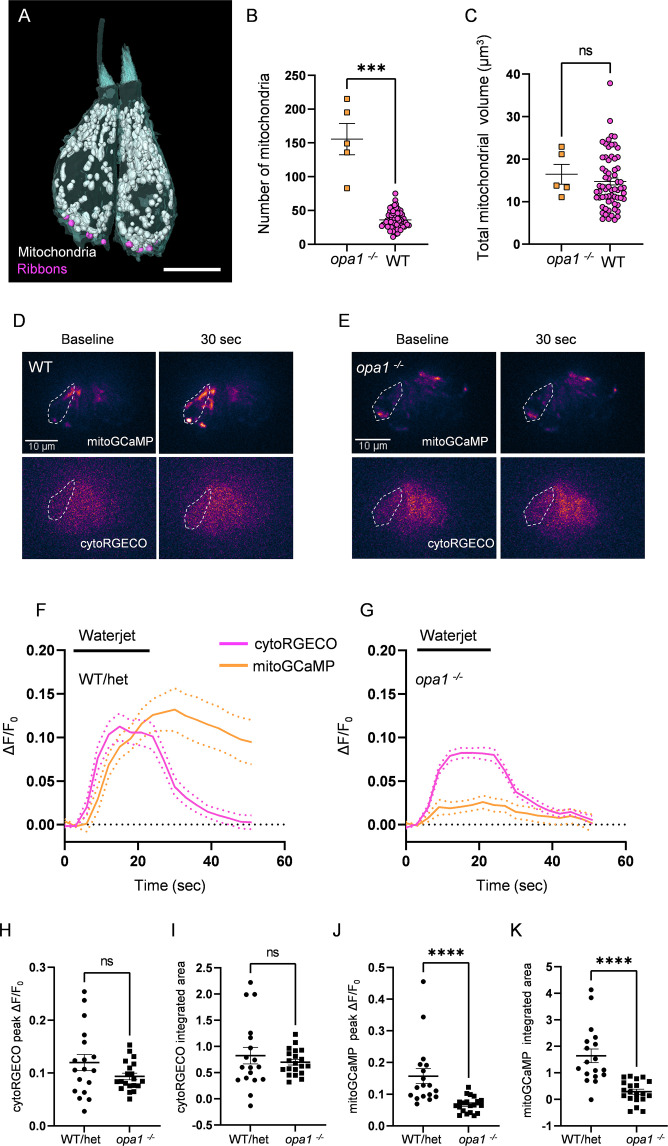
(A) Two representative opa1 HCs (Figure 5—source data 2). Scale bar = 5 µm. (B) Number of individual mitochondria in WT and opa1 HCs. WT: 36.1 ± 1.6; opa1: 155.6 ± 23.22. Kolmogorov–Smirnov test, p=0.0002. (C) Total mitochondrial volume in WT and opa1 HCs. In µm3, WT: 14.8 ± 0.8; opa1: 16.5 ± 2.3. Kolmogorov–Smirnov test, p=0.66. (B, C) opa1 HCs; n = 5 HCs, 1 NM, 1 fish. (D) Representative WT images of myo6:mitoGCaMP (top) and myo6:cytoRGECO (bottom) at baseline and following a waterjet stimulus. (E) Same as in (D), but for opa1 HCs. (F, G) Changes in myo6:mitoGCaMP and myo6:cytoRGECO Ca2+ signal (expressed as ΔF/F0) following a 20 s, 10 Hz waterjet for both WT/het (F) and opa1 HCs (G). (H) Peak myo6:cytoRGECO ΔF/F0 signal. WT/het: 0.1 ± 0.02; opa1: 0.1 ± 0.01, Mann–Whitney test, p=0.28. (I) Integrated myo6:cytoRGECO ΔF/F0 signal. WT/het: 0.8 ± 0.2; opa1: 0.7 ± 0.05, Mann–Whitney test, p=0.94. (J) Peak myo6:mitoGCaMP ΔF/F0 signal. WT/het: 0.15 ± 0.02; opa1: 0.05 ± 0.007, Mann–Whitney test, p<0.0001. (K) Integrated myo6:mitoGCaMP ΔF/F0 signal. WT/het: 1.6 ± 0.3; opa1: 0.2 ± 0.1, Mann–Whitney test, p<0.0001. (D–K) WT/het: n = 18 HCs, 7 fish. opa1: n = 20 HCs, 9 fish. Data are presented as the mean ± SEM.
Mutations in opa1 disrupt hair cell (HC) mitochondrial architecture and calcium buffering.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife