Fig. 1
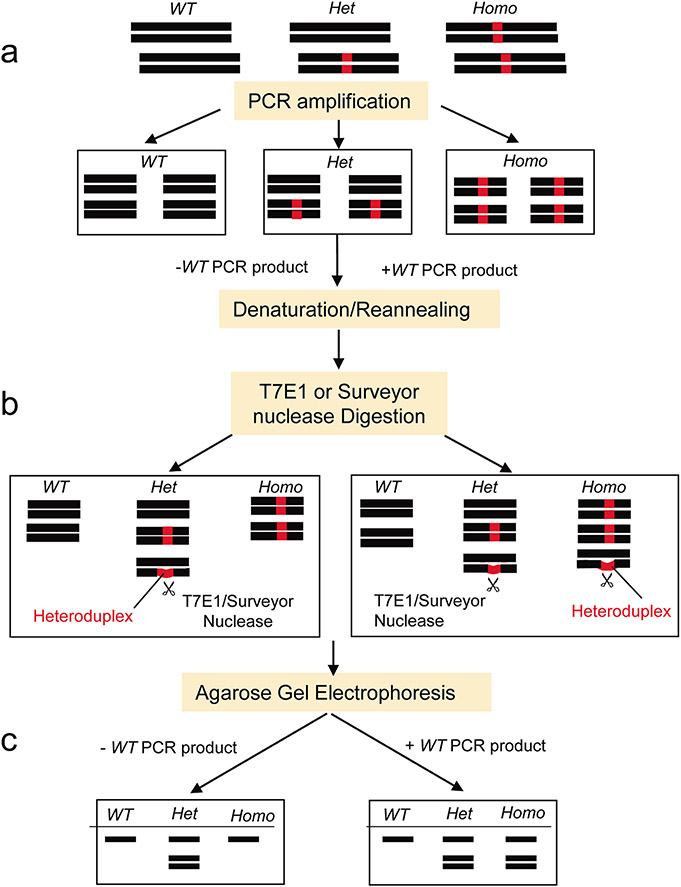
Genomic DNA was released from the samples following tissue lysis. The cell lysate was then directly used for the PCR to amplify the target gene region. The PCR product was split into two aliquots. In one aliquot, a wild-type (WT) control PCR product was added; in another aliquot, no WT control PCR product was added. The PCR products were then denatured at 95°C and reannealed. Heteroduplexes of the DNA fragments were expected to form in the heterozygous (Het) mutant products in this step. However, when mixed with a WT control PCR product, the artificial heteroduplexes of DNA were also expected to form in the homozygous (Homo) mutant samples, but not in the wild-type samples. T7 endonuclease I (T7E1) recognized and cleaved the heteroduplexes with a mismatch ≥2 base pairs but the Surveyor nuclease could cleave a single base mismatch and indels up to at least 12 nucleotides. Thus, we could clearly differentiate the WT, Het, and Homo animals with various sizes of mutations based on the cleaved products revealed on 2% agarose gel electrophoresis.