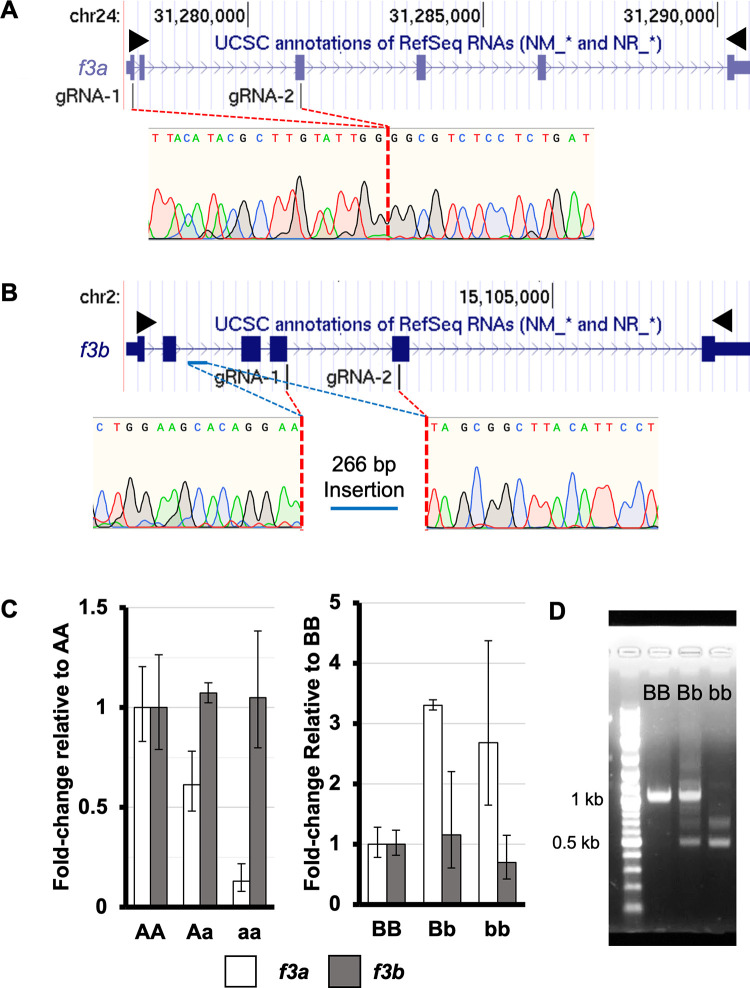
Fig 1
Two sgRNAs each for f3a and f3b were complexed with Cas9 and injected into single cell embryos. (A) A large deletion was identified between exon 1 and exon 3 in f3a resulting in a nonsense mutation. (B) A deletion was introduced between exon 4 and exon 5 of f3b. During endogenous repair, a section of intronic DNA (blue bar) was inverted and inserted at the site of the deletion introducing a premature stop codon. Black arrowheads indicate the locations of primers used for full length cDNA amplification. (C) RT-qPCR at 4 dpf demonstrated that the mutant f3a allele resulted in lower mRNA levels (consistent with nonsense mediated decay of the residual transcript) without affecting f3b levels. Conversely, loss of f3b induces upregulation of f3a, but the levels of f3b transcript were not diminished. (D) f3b cDNA was isolated from pools of 4 dpf larvae from heterozygous mutant incrosses. The expected wild-type band was visible in the BB and Bb mutants. Two smaller bands were evident in the Bb and bb pools, consistent with alternative splicing of the mutant transcript.