Figure 7.
The −1,995 bp sox2 enhancer element is required for HC regeneration
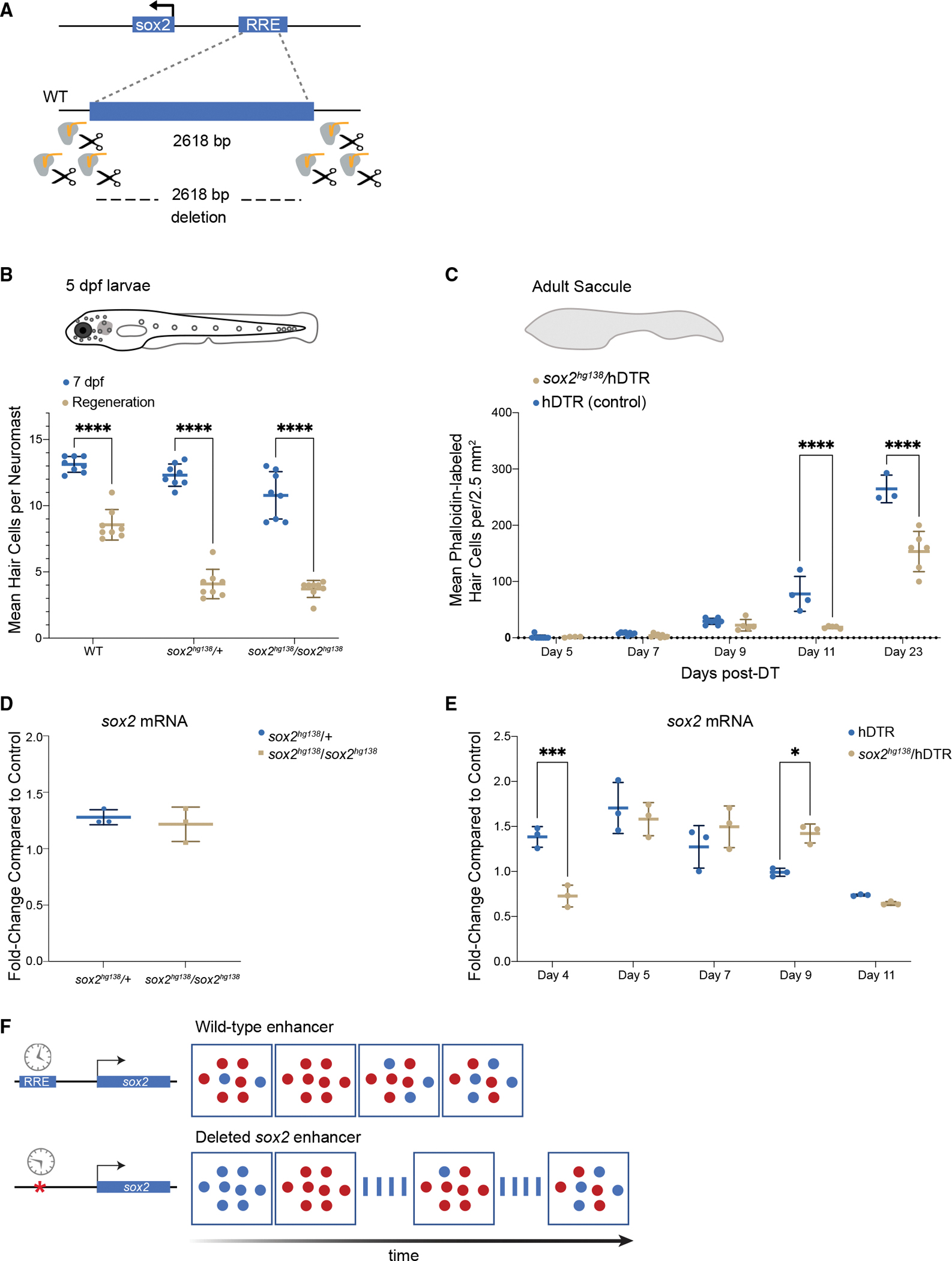
(A) Generation of enhancer deletion mutants using CRISPR-Cas9 editing.
(B) Lateral line HC regeneration is strongly inhibited 2 days after CuSO4 ablation in homozygous (soxhg138/soxhg138) and heterozygous (soxhg138/+) enhancer deletion mutants. The average number of HCs and SD are shown in the graph. A two-way ANOVA comparison and Sidak multiple comparison test of the data obtained on untreated larvae with the data obtained from CuSO4 treated larvae ∗∗∗∗p < 0.0001. Error bars show SD. n = 8 larvae in each group.
(C) Adult HC regeneration is significantly inhibited up to 23 days after HC ablation in heterozygous (soxhg138/hDTR) enhancer deletion mutants. The average number of HCs is shown in the graph. A two-way ANOVA comparison of the data obtained on regenerating control Tg(myo6b:hDTR) saccule (hDTR, blue) with the data obtained from regenerating heterozygous enhancer deletion mutant (soxhg138/hDTR, beige) saccule and Sidak multiple comparison test: ∗∗∗∗p < 0001. Error bars show SD. n = 6–8 saccules in each group unless otherwise indicated.
(D) Quantitative real-time PCR measuring sox2 mRNA levels in adult homeostatic sensory epithelia (saccule) from heterozygous (sox2hg138/+, blue) and homozygous (sox2hg138/sox2hg138, beige) enhancer deletion mutants. Fold-change compared with homeostatic sensory epithelia (saccule) of wild-type controls of the same age.
(E) Quantitative real-time PCR measuring sox2 mRNA levels in regenerating sensory epithelia (saccule) from Tg(myo6b:hDTR) controls (hDTR, blue) and sensory epithelia from heterozygous enhancer deletion mutants (sox2hg138/hDTR, beige) shows that activation of sox2 expression is delayed by 24 h (day 4) then remains elevated for an additional 24 h (day 9). The delay in sox2 expression at day 4 and the persistent expression at day 9 are statistically significant. A two-way ANOVA comparison of the data obtained on regenerating control (hDTR, blue) saccule with the data obtained from regenerating heterozygous enhancer deletion mutant saccule (soxhg138/hDTR, beige) and Sidak multiple comparison test: ∗p < 0.02, ∗∗∗p < 0.0004. Error bars show SD and triplicate technical replicates from dissected saccule of six to eight adult fish are shown in the graph.
(F) The upstream enhancer is involved in regulating the timing of sox2 expression but not essential for triggering activation. sox2 gene activation occurring in the supporting cells is depicted by the appearance of red dots. sox2 levels are delayed in enhancer deletion mutants compared with wild-type, but reaches the appropriate levels 24 h later. The return to baseline expression is also delayed by 24 h.