Fig. 6
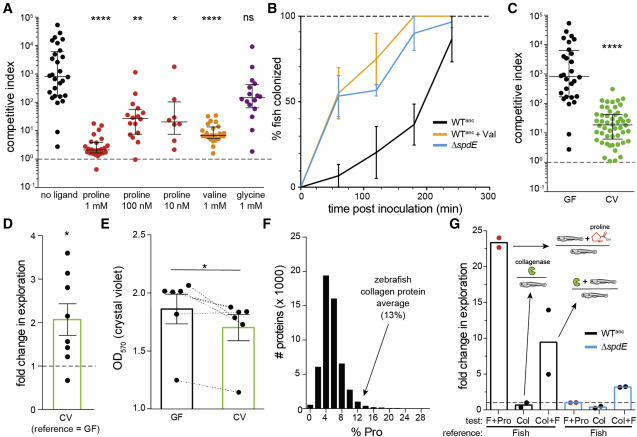
Figure 6. Aer01 host colonization is modulated by SpdE ligands and mediated by the microbiota (A) Competitive indices of ΔspdE (competed against the WT reference strain) ± added amino acids in the system. Dotted line indicates CI of 1 (i.e., no competitive advantage). n ≥ 8 fish/condition, 1–3 experimental replicates. Median and interquartile ranges are plotted. Statistical significance compared with no amino acid control group determined by Kruskal-Wallis with Dunn’s multiple comparisons, ∗∗∗∗p < 0.0001; ∗∗p < 0.01; ∗p < 0.05; ns, not significant. (B) Determination of the effect of SpdE ligand (Val, valine; 1 mM) on immigration rate of WTanc (see Figure S3A). The WTanc (no Val) and ΔspdE data are the same as plotted in Figure 2C; included for reference. n = 2–3 experimental replicates per condition. Means (± SEM) are plotted. (C) Competitive indices of ΔspdE (competed against the WT reference strain) in GF and CV larval zebrafish. Median and interquartile ranges are plotted. ∗∗∗∗p < 0.0001, two-tailed Mann-Whitney test. n ≥ 28 (GF) or 52 (CV) fish combined from 3 (GF) or 6 (CV) independent experiments. (D) Motility of WTanc determined by exploration assay (see Figure S7) comparing motility in CV fish-conditioned flask media (FC-FM) to GF FC-FM. Each data point represents an independent experiment. n = 8 independent experiments using media collected from different flasks of GF and CV larval zebrafish. Bar, mean (± SD). Dotted line represents fold change of 1 (i.e., no difference). ∗p < 0.05; one-sample t test, statistically different than 1. (E) Quantification of biofilm formation for WTanc in GF and CV FC-FM. Dotted lines connect data comparing GF and CV FC-FM collected on the same day from fish from the same egg clutch. n = 6 experimental replicates; each data point represents an independent experiment using CV or GF FC-FM collected from a different fish flask. ∗p < 0.05, paired t test. (F) Histogram of the number of predicted proteins in the zebrafish genome, binned according to % proline. Pro, proline. (G) Quantification of WTanc (black outlined bars) and ΔspdE (blue outlined bars) motility (exploration assay) in supernatant from collagenase-digested larval zebrafish (Col + F). Supernatant from untreated fish (Fish), untreated fish spiked with proline (F + Pro), and a collagenase only (Col) are included as references. Each data point represents an independent experiment; “reference” strain/condition indicated on bottom. n = 2 experimental replicates. Bar, mean. Dotted line represents fold change of 1 (i.e., no difference). See also Figure S7.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Host Microbe