Fig. 3
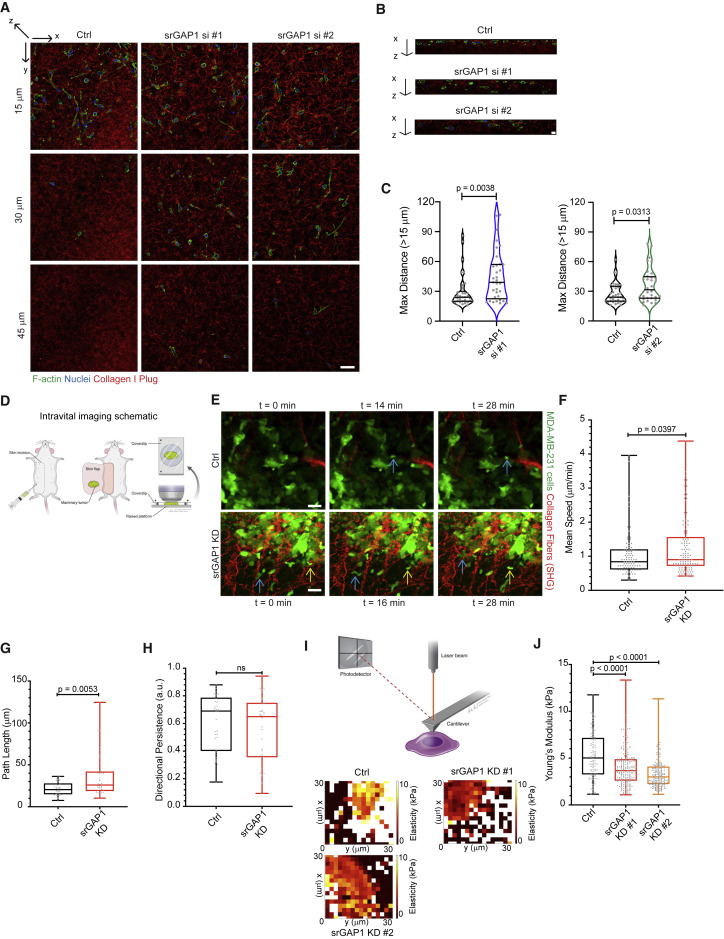
Figure 3. In vitro and in vivo invasion are regulated by srGAP1 expression (A) Representative images of Ctrl, srGAP1 si #1, and srGAP1 si #2 MDA-MB-231 cells 3 days after being seeded on top of collagen I plugs. Images taken at 15, 30, and 45 μm in the z direction (deeper into collagen plug away from surface). Cells are labeled with phalloidin and DAPI, collagen I is fluorescently labeled. Scale bar, 50 μm. (B) x-z images of representative collagen I plugs from (A). Scale bar, 10 μm. (C) Maximum distance (>15 μm) Ctrl and srGAP1 si #1 cells (left) or Ctrl and srGAP1 si #2 MDA-MB-231 cells (right) invade into the collagen I plug. Each value is a single cell/field. Left: n = 37 (Ctrl), 34 (srGAP1 si #1) cells. Right: n = 38 (Ctrl), 35 (srGAP1 si #2) cells. Data pooled over five independent experiments and represented as a violin plot; horizontal lines are median and interquartile range. Mann-Whitney U test, two-tailed. (D) Schematic of intravital imaging setup. (E) Three individual time frames from intravital imaging movies of Ctrl and srGAP1 KD #1 MDA-MB-231 primary tumors. Tumor cells in green, collagen fibers in red through second harmonic generation (SHG). Blue and yellow arrows indicate in vivo cell motility over time. One z slice shown at 0, 16, 28 min. Scale bar, 25 μm. (F) Average mean speed of tumor cells moving in μm/min in vivo from (C). Data pooled from 3 Ctrl and 3 srGAP1 KD #1 mice, n = 94 (Ctrl) and 102 (srGAP1 KD #1) MDA-MB-231 tumor cells. Mann-Whitney U test, two-tailed. Data represented as a box and whisker plot, with median and interquartile range in the box and min and max as whiskers. (G) Average path length of tumor cells that appear in a minimum of 10 frames of intravital imaging movies (t = 20–28 min). Data pooled from 3 Ctrl and 3 srGAP1 KD #1 mice, n = 27 (Ctrl) and 33 (srGAP1 KD #1) MDA-MB-231 tumor cells. Mann-Whitney U test, two-tailed. Data represented as a box and whisker plot, with median and interquartile range in the box and min and max as whiskers. (H) Directional persistence of tumor cells that appear in a minimum of 10 frames of intravital imaging movies (t = 20–28 min). Data pooled from 3 Ctrl and 3 srGAP1 KD #1 mice, n = 27 (Ctrl) and 33 (srGAP1 KD #1) MDA-MB-231 tumor cells. Mann-Whitney U test, two-tailed. Data represented as a box and whisker plot, with median and interquartile range in the box and min and max as whiskers. (I) Top: schematic of atomic force microscopy setup. Bottom: elasticity heatmaps of representative Ctrl, srGAP1 KD #1, and srGAP1 KD #2 MDA-MB-231 cells. (J) Modulus of elasticity of Ctrl, srGAP1 KD #1, and srGAP1 KD #2 MDA-MB-231 cells using AFM. n = 100 (Ctrl), 200 (KD #1), and 200 (KD #2) measurements over 4, 8, and 8 cells, respectively. Mann-Whitney U test, two-tailed. Data represented as a box and whisker plot, with median and interquartile range in the box and min and max as whiskers.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Rep.