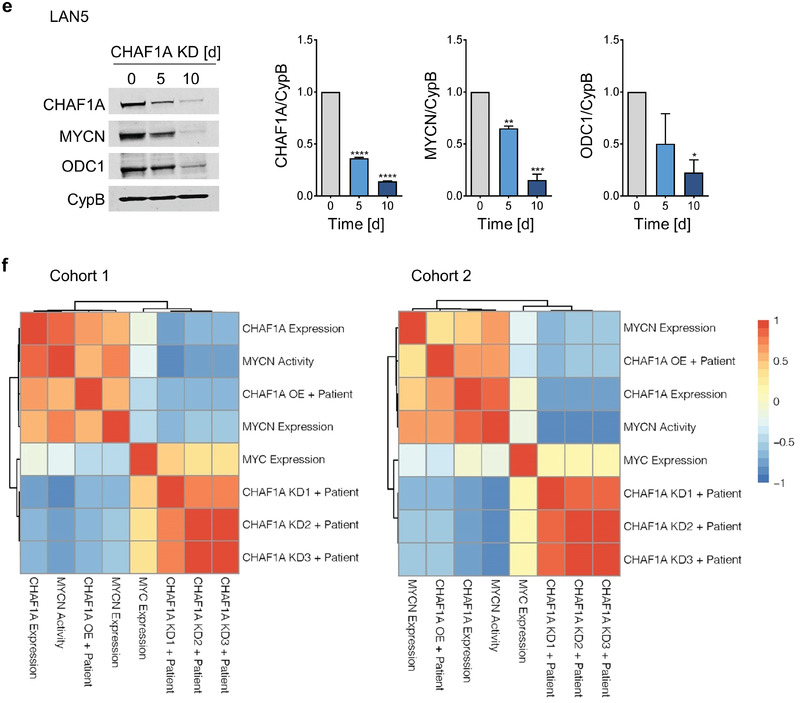
Figure 6 (continued)
CHAF1A is a direct target of MYCN. a,b) mRNA and protein expression of CHAF1A in TET‐21/N cells when MYCN is turned off upon DOX treatment (2 µg mL−1, 24–96 h). GAPDH is used as housekeeping gene, CypB as protein loading control. Data are mean ± SD (