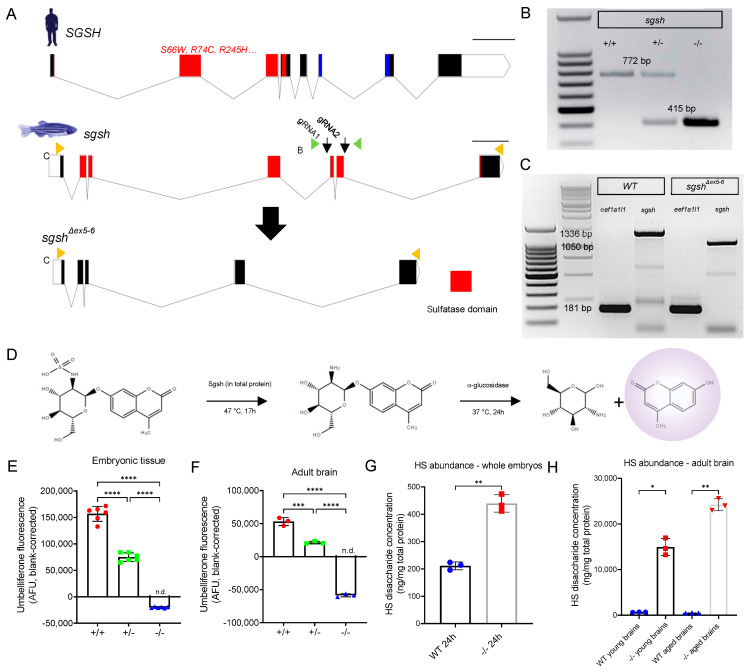
Figure 1 Generation and biochemical validation of the sgshΔex5−6 mutant zebrafish. (A) Exon-intron diagram representations of wild type human SGSH, wild type zebrafish sgsh, and the zebrafish sgshΔex5−6 allele. Exons comprising sulfatase domain highlighted in red; exons comprising sulfatase-associated Domain of Unknown Function DUF4976 highlighted in blue. Downwards-facing arrows indicate gRNA target sites. Orange arrowheads reflect primer positions for RT-PCR; green arrowheads reflect primer positions for genotyping PCR. Scale bar is 1 kilobase. (B) Genotyping PCR of wild type (+/+), heterozygous (+/−) and homozygous sgshΔex5−6 (−/−) alleles. Wild type sgsh produces a band at 772 bp, while sgshΔex5−6 produces a band at 415 bp. (C) RT-PCR amplicons of sgsh in RNA from wild type (+/+) or homozygous sgshΔex5−6 (−/−) zebrafish embryos at 24 hpf using primers located in the first and last exons of sgsh. Wild type sgsh produces a prominent band at 1336 bp, while sgshΔex5−6 produces a band at 1050 bp corresponding to loss of exons 5 and 6. eef1a1l1 (amplicon size 181 bp) included for both genotypes as housekeeping control. (D) Overview of Sgsh enzyme activity assay. 4MU-αGlcNS is desulfated by Sgsh in total protein, then glucosamine is hydrolysed from UV-fluorescent 4MU by bacterial α-glucosidase. (E) Enzyme activity in wild type (+/+), sgshΔex5−6 heterozygous (+/−) and homozygous (−/−) whole embryos (n = 100 embryos per sample, 6 samples per group). (F) Enzyme activity in adult brain (n = 3 brains per sample, 3 samples per group) for each genotype. Negative arbitrary fluorescence values observed in sgshΔex5−6 homozygous samples result from absorbance of UV excitation wavelengths by protein input and blank correction. Data in (E,F) are presented as mean ± SEM and tested via ordinary one-way ANOVA with Tukey’s multiple comparisons test; **** p < 0.0001 and *** p = 0.0001. (G) HS disaccharide abundance in whole 24 hpf embryos from either wild type or sgshΔex5−6 genotypes as determined by LC-MS/MS (n = 50 embryos per sample, n = 3 samples per group). Data presented as mean ± SEM and tested by Welch’s t-test. ** p = 0.0021. (H) HS abundance in young (6-month-old) or aged (18-month-old) brains from either wild type or sgshΔex5−6 genotypes determined by LC-MS/MS (n = 3 brains per sample, n = 3 samples per group). Data presented as mean ± SEM and tested by Brown-Forsythe and Welch ANOVA with Dunnett’s T3 multiple comparisons test. * p = 0.0171, ** p = 0.0031.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Int. J. Mol. Sci.