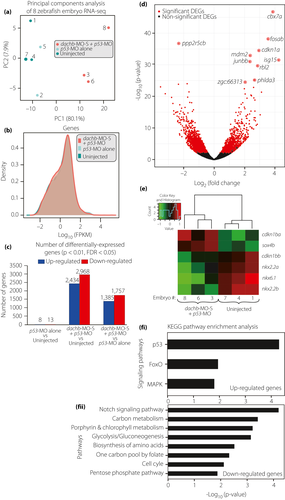
Fig. 9 dachb knockdown changed the messenger ribonucleic acid (RNA) transcriptome profile of zebrafish embryos at 30 h post-fertilization. (a) Principal components analysis of the RNA sequencing (RNA-Seq) datasets. Each data point represents a zebrafish RNA-Seq sample and the color represents the sample group: splice-blocking dachb-morpholino (dachb-MO-S) + p53-MO (red), p53-MO alone (light cyan) and uninjected control (dark cyan). (b) Density distribution plots of log expression level (fragments per kilobase of transcript per million) for all genes in the dachb-MO-S + p53-MO (red), p53-MO alone (light cyan) and uninjected control (dark cyan) groups. (c) Number of differentially expressed genes (P < 0.01, false discovery rate <0.05) in the three comparison groups. (d) Volcano plot of log2(Fold Change) versus –log10(P-value) for all 27,957 genes in the dachb-MO-S + p53-MO versus p53-MO alone groups. Differentially expressed genes are in red (P < 0.01, false discovery rate <0.05). The 10 most significant differentially expressed genes in terms of P-value are labeled. (e) Comparison of the level of expression of genes known to be involved in pancreas development after injection with dachb-MO-S + p53-MO, compared with uninjected controls. (f) Significantly enriched KEGG pathways (P < 0.05, false discovery rate <0.2) of (fi) up- and (fii) downregulated genes in the dachb-MO-S + p53-MO versus p53-MO alone groups.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ J Diabetes Investig