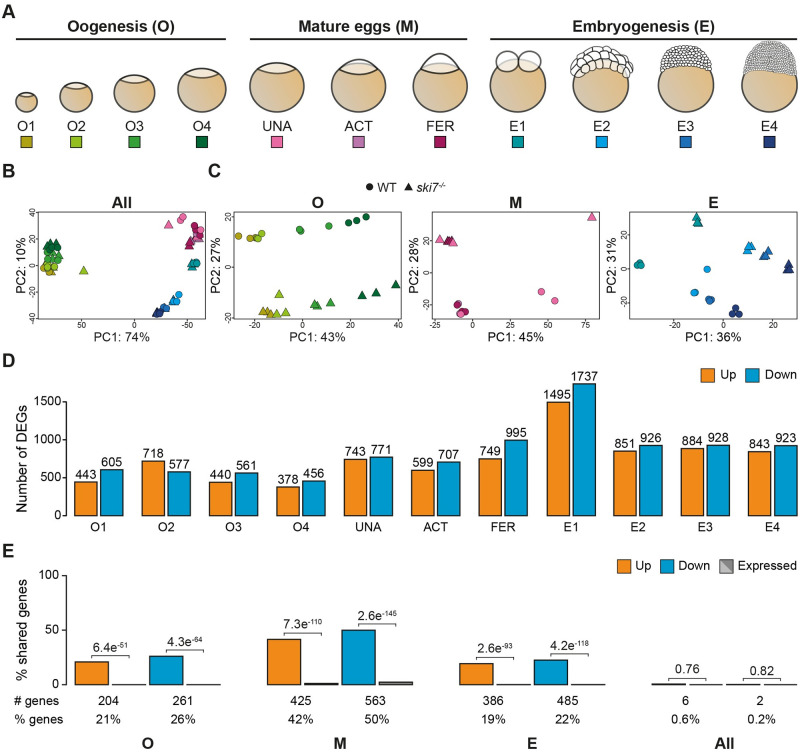
Fig 3
(A) Schematic representation of the stages used for RNA-seq during three consecutive developmental periods: oogenesis, mature eggs and embryogenesis. (B, C) Principal Component Analyses (PCA) of all time points (B) or individual periods (C) used for the polyA+ RNA-seq comparison of