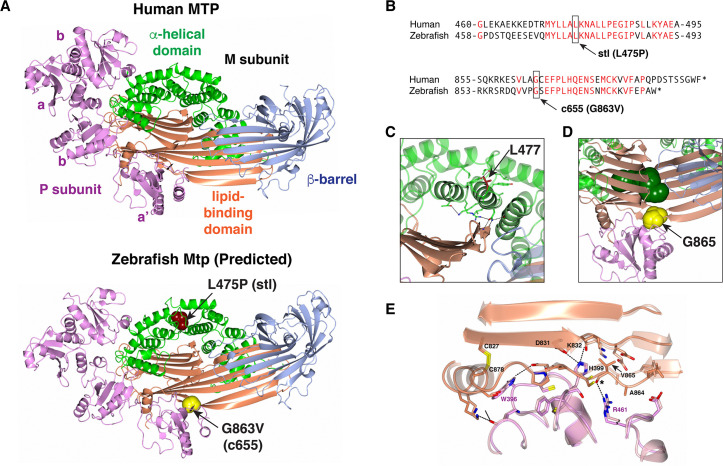
Fig 7
(A) Ribbon representation of the human MTP complex (PDB entry 6I7S) and the Zebrafish modeled structure. The positions of L475 and G863 in the Zebrafish structure are shown in space-filling representation. (B) Alignment of human MTP and zebrafish Mtp amino acid sequences surrounding the