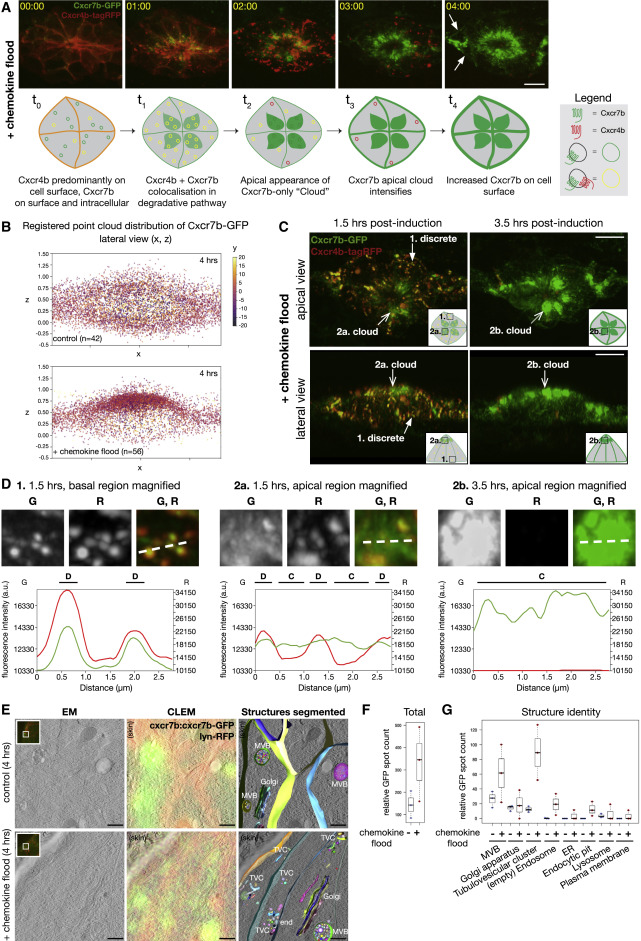
Fig. 4 Cxcr7b Upregulation Is Characterized by a Degradation-to-Recycling Trafficking Switch (A) Time lapse of Cxcr7b-GFP and Cxcr4b-tagRFP in chemokine flood (top). Arrows: Cxcr7b-GFP on the PM. Schematic of Cxcr7b-GFP and Cxcr4b-tagRFP trafficking itineraries in chemokine flood (bottom). Scale bar, 10 μm. See Video S4. (B) Registered point cloud 2D scatterplots of Cxcr7b-GFP intensity distributions in control (top) and chemokine flood (bottom). n = 42 and 56. (C) Cxcr7b-GFP/Cxcr4b-tagRFP after indicated times post-induction of chemokine flood. Apical (top) and lateral (bottom) views. Closed arrows, discrete signals and open arrows, non-discrete cloud-like signals. Scale bar, 10 μm. (D) Intensity line profiles to compare trafficking behaviors of Cxcr7b-GFP and Cxcr4b-tagRFP from the indicated regions and times. D, discrete and C, cloud. (E) CLEM of Cxcr7b-GFP and krt15:lyn-RFP in control (top) or chemokine flood (bottom). From left: EM, single slice from an EM tomogram; CLEM, overlay of EM with on section light microscopy (LM); segmented structures, overlay of segmented GFP-positive organelles; and RFP-positive PM with EM. Insets, on-section LM and boxes, targeted regions where EM was acquired. G, Golgi apparatus; MVB, multivesicular body; TVC, tubulovesicular cluster; end, endocytic pit. Scale bars, 1 μm. See Video S5. (F) Relative GFP spot counts as total divided by tomogram volume. Biological repeats = 3. Number of structures: control n = 85, 98, and 43 and chemokine flood n = 31, 128, and 175. (G) Relative GFP spot counts per organelle category.
Reprinted from Developmental Cell, 52(4), Wong, M., Newton, L.R., Hartmann, J., Hennrich, M.L., Wachsmuth, M., Ronchi, P., Guzmán-Herrera, A., Schwab, Y., Gavin, A.C., Gilmour, D., Dynamic Buffering of Extracellular Chemokine by a Dedicated Scavenger Pathway Enables Robust Adaptation during Directed Tissue Migration, 492-508.e10, Copyright (2020) with permission from Elsevier. Full text @ Dev. Cell