Fig. 1
slc6a3 Deficiency Causes Repetitive Behavior
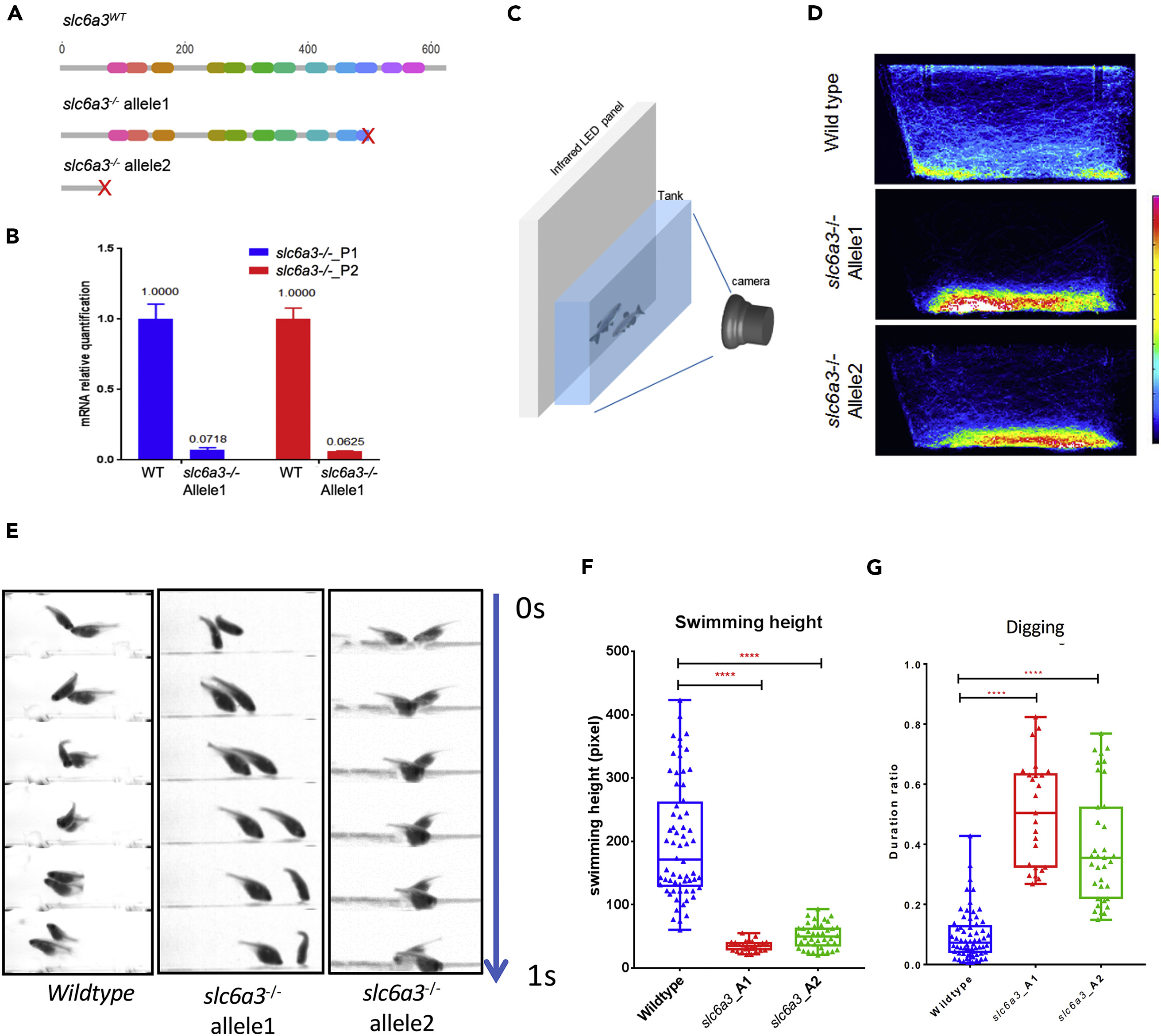
(A) The wild-type Slc6a3 protein structure and two different alleles obtained by CRISPR deletions resulting in truncation at different domains in Slc6a3 proteins.
(B) qPCR indicating dramatic reduction of slc6a3 truncated mRNA in slc6a3−/− allele1 when compared with the wild-type.
(C) The behavior recording system.
(D) Heatmaps of fish trajectory of wild-type and two alleles of slc6a3−/− in the home tank in a 30-min video (60 frames per second). The heatmap colors represent frames that fish appear within each 5 × 5-pixel bin in the image (1224 × 500 pixel in 30 min).
(E) The “digging” behavior in the slc6a3−/− fish compared with wild-type. All 1-s videos show fish at the tank bottom. Fish bearing both slc6a3−/− alleles display a persistent “digging” behavior, whereas wild-type fish do not.
(F) The quantification of average swimming height of fish (nWT = 64, nslc6a3_A1 = 25, nslc6a3_A2 = 40, ****p < 0.0001, significance test: one-way ANOVA Kruskal-Wallis test).
(G) The quantification of duration of “digging” feature over the whole 30-min recording time in the tank using Janelia Automatic Animal Behavior Annotator (JABBA) (Kabra et al., 2013) (nWT = 64, nslc6a3_A1 = 25, nslc6a3_A2 = 40, ****p < 0.0001, significance test: one-way ANOVA Kruskal-Wallis test.)
See Figure S3 for more details.