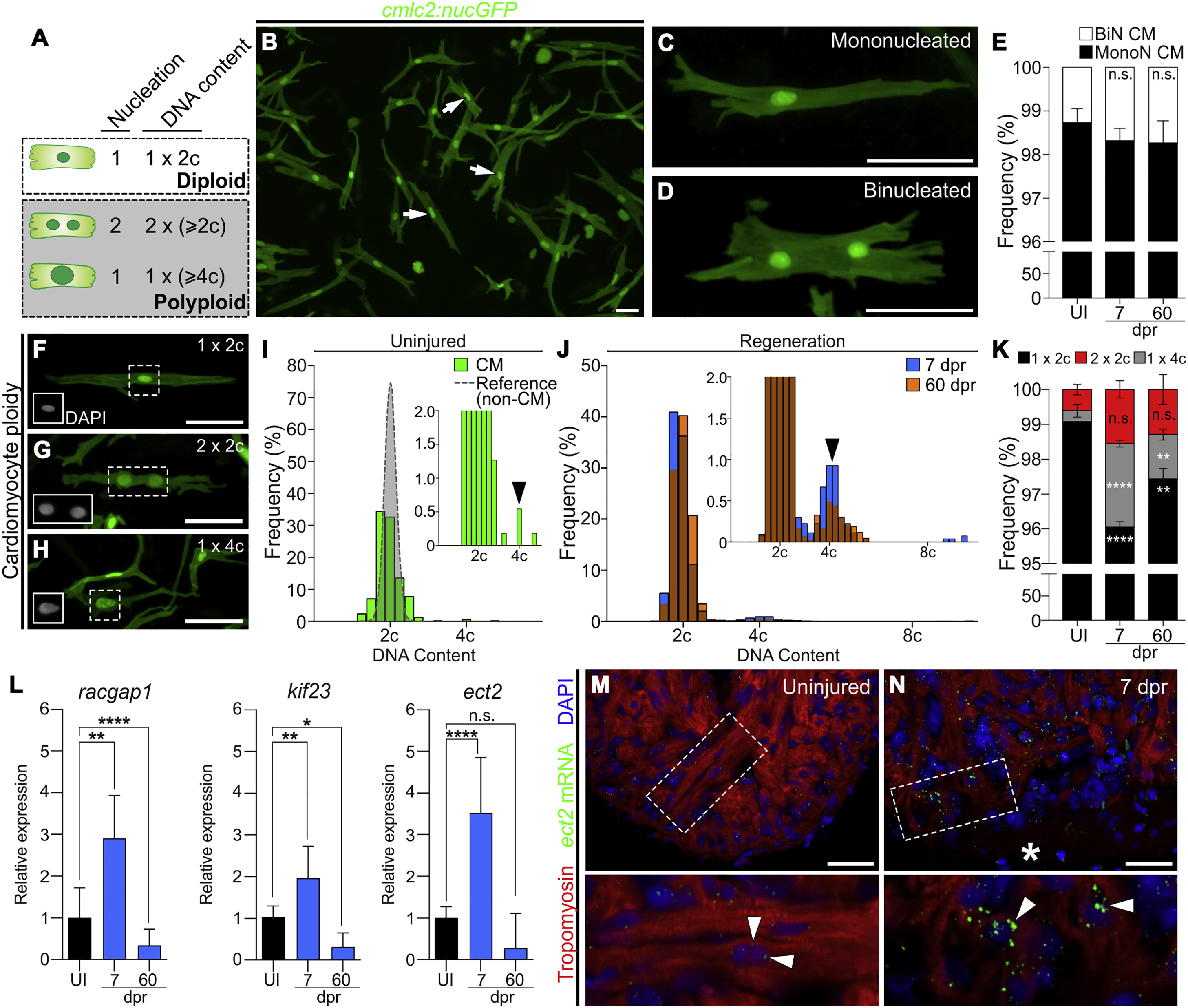
Fig. 1
Zebrafish Cardiomyocytes Are Mononucleated, Diploid, and Upregulate Ect2 during Heart Regeneration
(A) Schematic depicting cardiomyocyte nucleation and ploidy. See also Figure S1.
(B) Cardiomyocytes (white arrows) from dissociated Tg(cmlc2:nucGFP) hearts.
(C and D) Mononucleated and binucleated Tg(cmlc2:nucGFP) cardiomyocytes.
(E) Average percentages of mononucleated (MonoN) and binucleated (BiN) cardiomyocytes in uninjured (UI), 7-dpr, and 60-dpr ventricles (mean ± SD, n = 9,309, 9,269, and 9,578 total cells, respectively, from 4 biological replicates per group; 2 pooled ventricles per replicate; n.s., not significant by one-way ANOVA test).
(F–H) DAPI-stained mononucleated diploid, binucleated tetraploid, and mononucleated tetraploid cardiomyocytes isolated from Tg(cmlc2:nucGFP) hearts. Insets show DAPI signal.
(I) Distributions of non-cardiomyocyte (gray) and cardiomyocyte (green) DNA content in homeostatic ventricles (n = 779 and 552 cells, respectively, from 3 biological replicates; 1 ventricle per replicate). Inset is magnification to show low-frequency events (black arrowhead indicates tetraploid cardiomyocytes).
(J) Distributions of cardiomyocyte DNA content in 7-dpr and 60-dpr ventricles (blue and orange bars, respectively; overlap appears in brown). Inset is magnification to show low-frequency events (black arrowhead indicates tetraploid cardiomyocytes).
(K) Quantification of indicated cardiomyocyte populations from UI, 7-dpr, and 60-dpr ventricles (mean ± SD, n = 3,250, 2,699, and 4,306 total cells from 7, 3, and 4 biological replicates per group, respectively; 3 ventricles per replicate; ∗∗∗∗p < 0.0001; ∗∗p < 0.01 by one-way ANOVA followed by Tukey's multiple comparisons test).
(L) qPCR analysis showing relative expression of three genes involved in cytokinesis in UI, 7-dpr, and 60-dpr ventricles (mean ± SD, n = 3 technical replicates, 10 biological replicates, 1 ventricle per replicate, ∗∗∗∗p < 0.0001; ∗∗p < 0.01; ∗p < 0.05 by one-way ANOVA followed by Tukey's multiple comparisons test). See also Figure S2.
(M and N) Single confocal sections of ect2 RNAScope (arrowheads) in UI and 7-dpr hearts. Boxed regions are shown at higher magnifications. n = 4 hearts per group with 3 sections per heart. Asterisk indicates wound edge.
CM, cardiomyocytes. Scale bars, 50 μm.
Reprinted from Developmental Cell, 44, González-Rosa, J.M., Sharpe, M., Field, D., Soonpaa, M.H., Field, L.J., Burns, C.E., Burns, C.G., Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish, 433-446.e7, Copyright (2018) with permission from Elsevier. Full text @ Dev. Cell