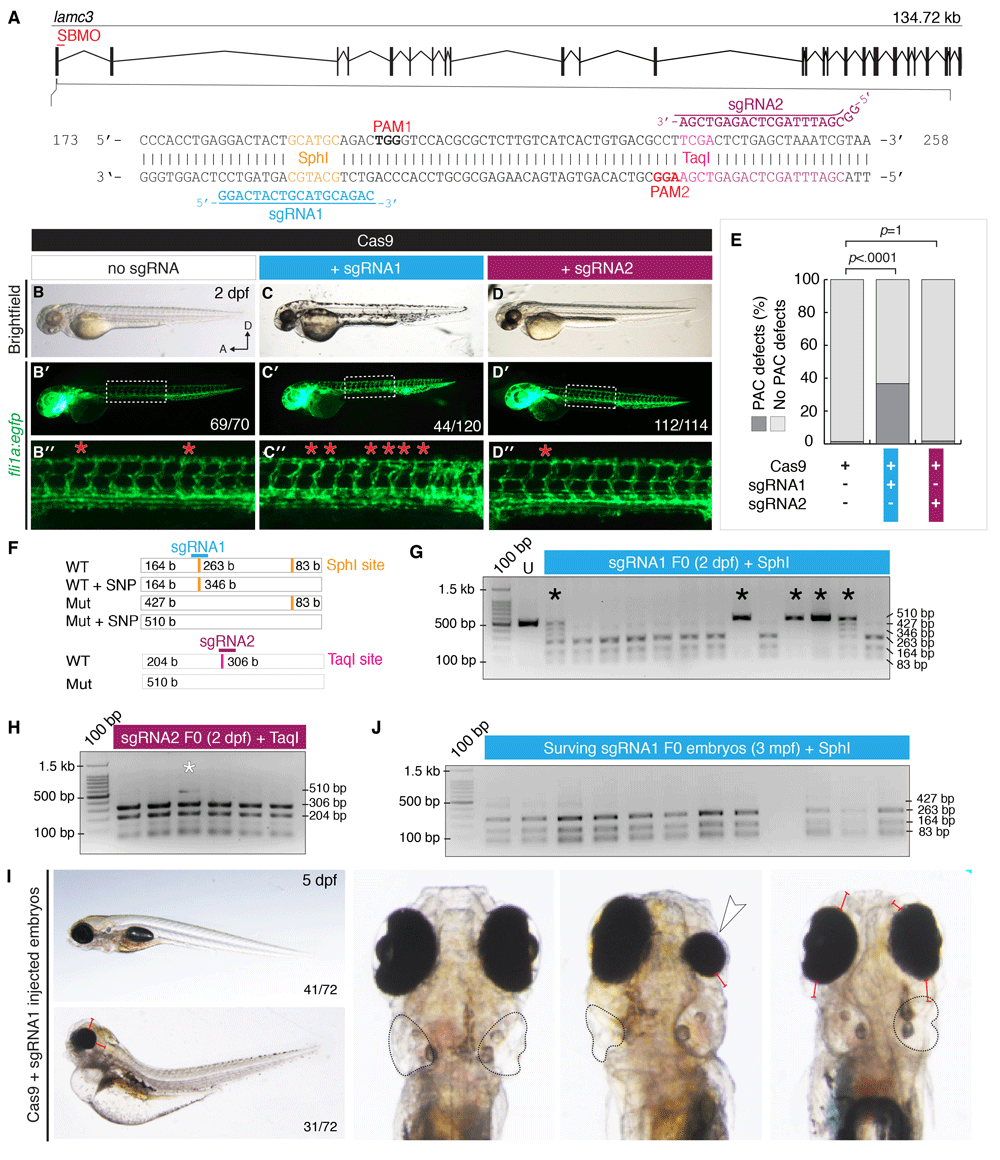
Fig. 3
CRISPR/Cas9 mutagenesis of lamc3 causes defects in parachordal chain (PAC) development.
(A) Design of sgRNAs targeted to exon1 of the lamc3 gene. Each sgRNA targets a restriction endonuclease site used for genotyping and is upstream of a protospacer adjustment motif (PAM). (B–D) Cas9/sgRNA injected embryos (as labelled) at 50 hpf. (B–D) Brightfield images show no developmental defects or developmental delay. (B′–D′) Whole embryo images of Tg(fli1a:egfp) fluorescence area indicated by white dotted line is enlarged in (B′′–D′′) the PAC in each hemisegment is indicated with a red asterisk. (E) Quantification of Tg(fli1a:egfp) embryos at 50 hpf with PAC defects. P-values determined by Fisher’s exact Two-tailed test compared to Cas9 alone controls. (F–I) 2% agarose genotyping gels with 100 bp ladder. (F) Schematic diagram of the region amplified by PCR contains two SphI sites (orange), which would digest into three fragments of 83, 164 and 263 bp in length. Predicted mutation of o