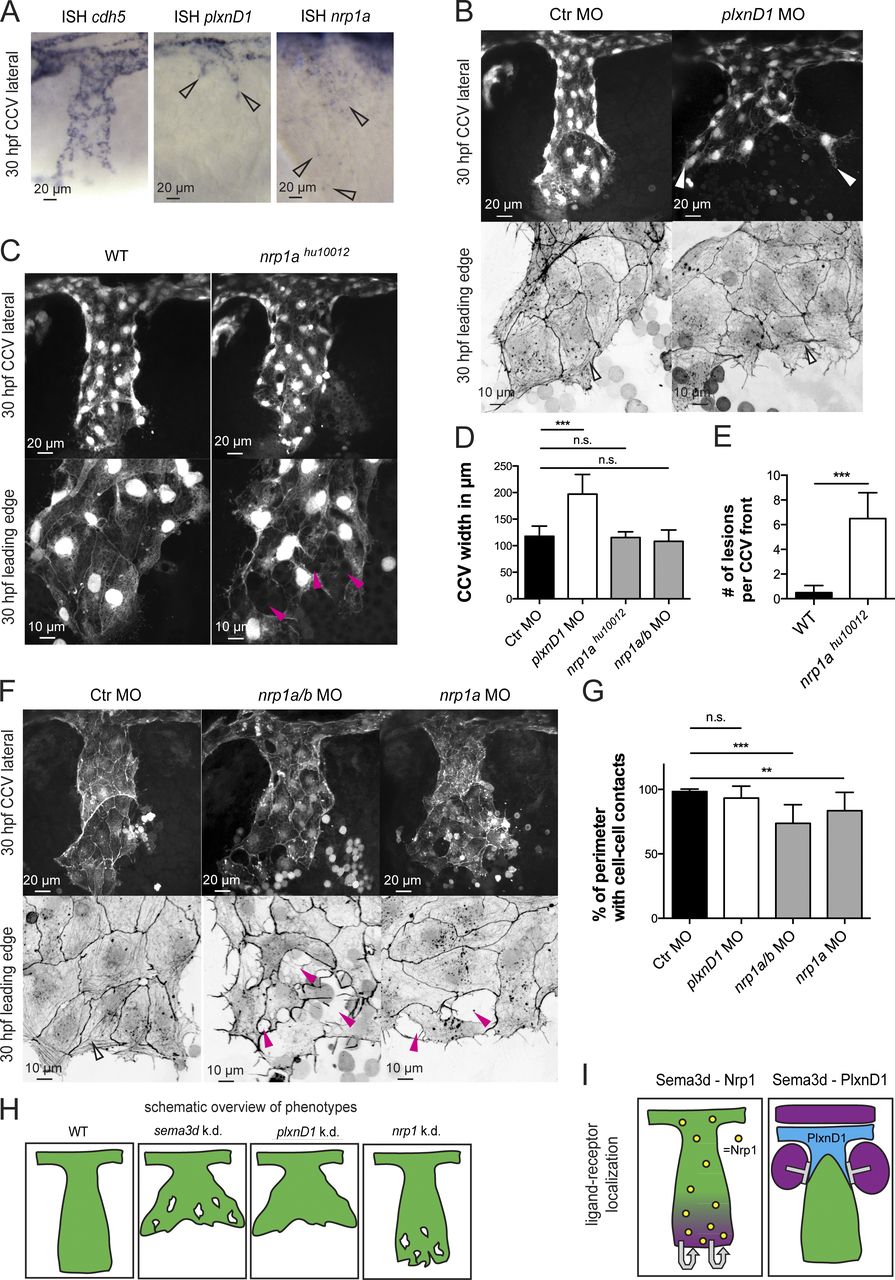
Fig. 7 Differential Sema3d functions are transduced by signaling through the different receptors Nrp1 or PlxnD1. (A) In situ hybridization (ISH) of 30-hpf embryos showing cdh5 expression in all CCV cells, plxnD1 expression in the dorsal-most CCV cells (open arrowheads), and nrp1 expression in all CCV cells (open arrowheads). (B–G) Analysis of PlxnD1 or Nrp1 deficiency in Tg(kdrl:EGFP)s843 or Tg(fli1a:lifeactEGFP)mu240 embryos at 30 hpf. (B) Loss of PlxnD1 led to a wider CCV (white arrowheads), but cell–cell contacts were not impaired, and parallel-arranged Actin cables were present (open arrowheads). (C) nrp1ahu10012 mutant embryos had reduced cell–cell contacts (pink arrowheads) but did not exhibit a wider CCV. (D) Quantification of CCV front width (each n = 11; nrp1ahu10012, n = 4). (E) Number of lesions in nrp1ahu10012 mutant embryos is increased compared with WT (n = 4). (F) nrp1a/b and nrp1a morphant CCVs exhibit a normal width, but the leading edges showed lesions (pink arrowheads) and lacked Actin cables (compare with open arrowhead in Ctr). (G) Quantification of cell–cell contact length (each n = 13). (H) Schematic overview of phenotypes comparing Sema3d deficiency with plxnD1 and nrp1 knockdown. (I) Schematic illustration of ligand-receptor localization indicating autocrine Sema3d-Nrp1 signaling and paracrine Sema3d-PlxnD1 signaling. Magenta, sema3d expression; green, CCV ECs; blue, plxnD1 expression; and yellow, nrp1 expression. ***, P < 0.001; **, P < 0.01; n.s., not significant. Error bars indicate SD.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ J. Cell Biol.