Fig. S3
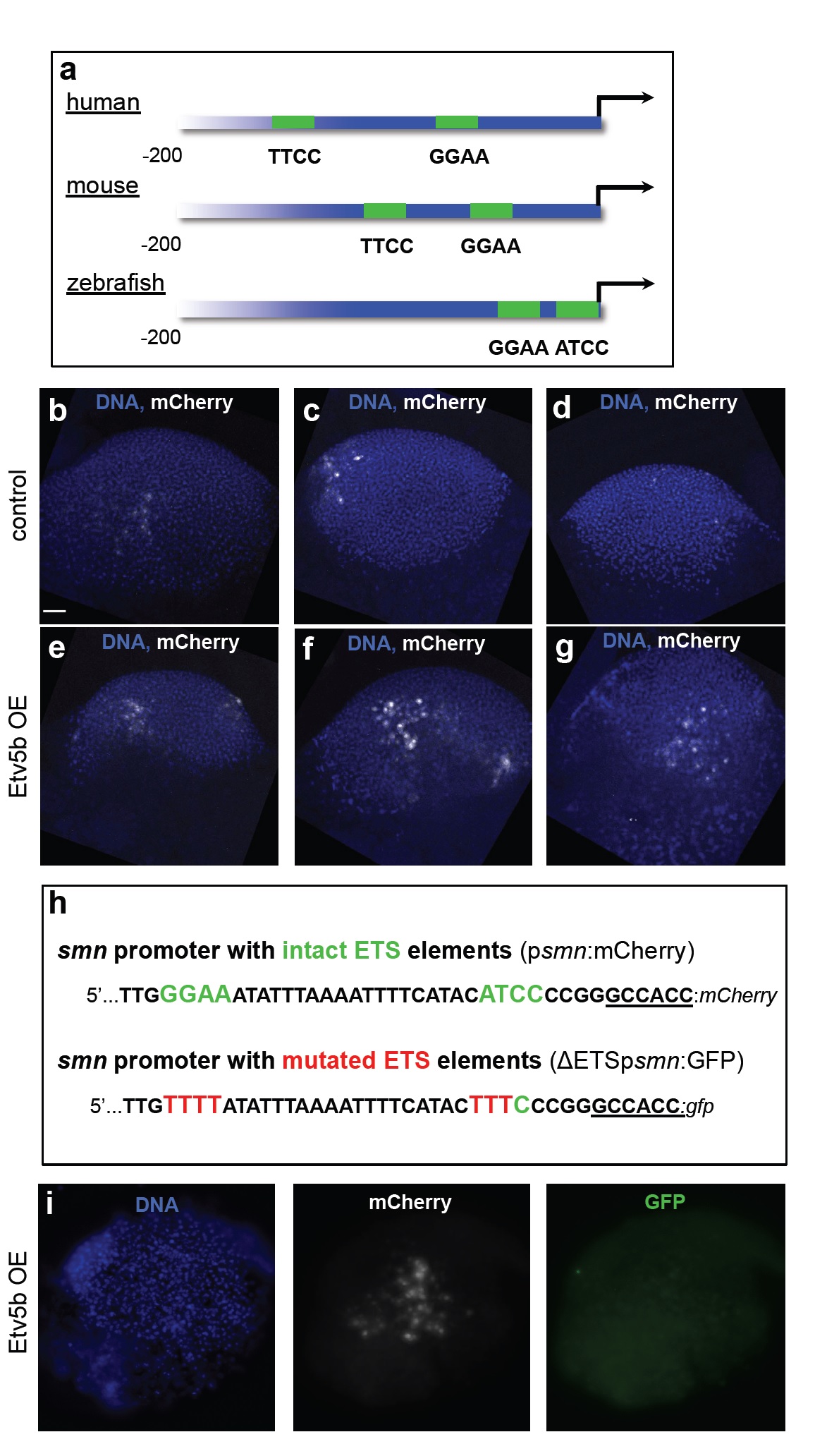
Evolutionary conservation of proximal ETS-binding sites in the smn promoter and overexpression and knock-down of etv5b.
a The two proximal ETS binding sites (with consensus sites being GGAA/T or A/TTCC) shown in green in the human, murine and zebrafish smn promoters. Note that the binding sites in human and mouse are on the forward strands, while in zebrafish it is located in the reverse strand. The first 200 nucleotides upstream of the start codons are shown.
b-g mCherry immunostaining (white) at the gastrula stage following psmn:mCherry injection without (b-d) or with Etv5b overexpression (e-g) on the same batch of embryos that was used for RT-PCR analysis shown in Fig. 3e. DNA is shown in blue. Scale bar: 100 microns. 8 embryos of control and 5 embryos with Etv5b overexpression were analyzed.
h Schematic representation of the first 40 nucleotides of the smn promoter with intact (psmn:mCherry, top) and mutated (ΔETSpsmn:GFP, bottom) ETS elements used in the experiment in panel i.
i A representative gastrula stage embryo that had been co-injected with Etv5b mRNA, the psmn:mCherry plasmid as well as the ΔETSpsmn:GFP plasmid and immunostained against mCherry and GFP. DNA is shown in blue. 5 embryos showing strong mCherry staining were analyzed.