Fig. 5
Itgα5 Conformation Regulates Itgα5 Clustering and FN Matrix Assembly in the Presomitic Mesoderm
Itgα5GAAKR contains two amino acid substitutions that bias toward the extended, active conformation.
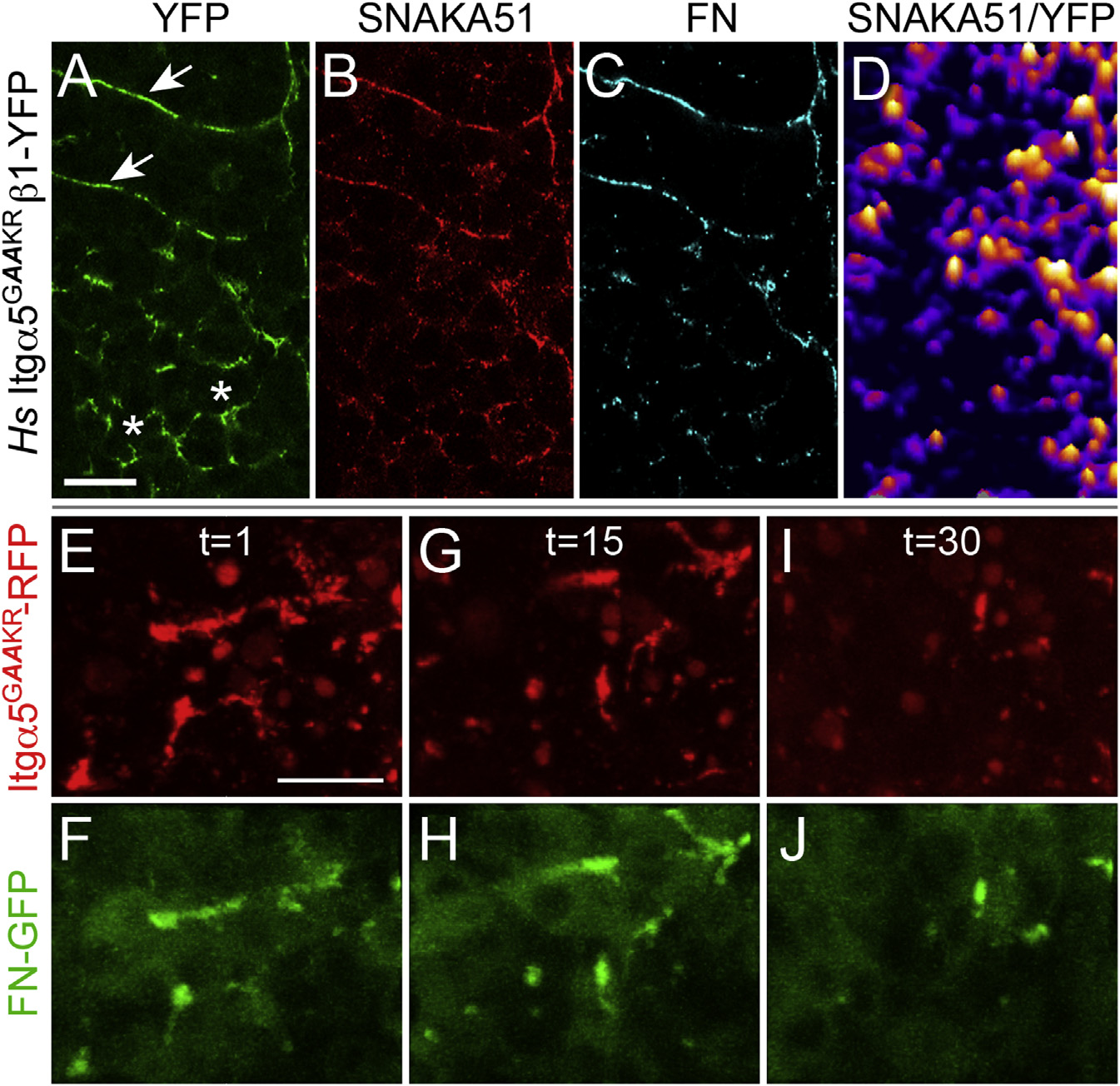
(A–D) Hs Itgα5GAAKRβ1-YFP (A) clusters along somite boundaries (arrows) in MZ itgα5-/- embryos, but also inappropriately clusters in the PSM (asterisks). This inappropriate clustering colocalizes with Hs Itgα5GAAKRβ1-YFP adoption of the active conformation (B and D) and FN aggregation (C). SNAKA51 and FN IHC were examined in 51 Hs Itgα5GAAKRβ1-YFP expressing MZ itgα5-/- embryos (five experiments).
(E–J) Hs Itgα5GAAKRβ1-RFP clustering and FN-GFP aggregation in the PSM is unstable. The time-lapses of three Tg(tbx6l:fn1a-GFP) embryos coinjected with the itgα5 morpholino and itgα5GAAKR-RFP mRNA were acquired and analyzed. The three time points t = 1 (E and F), t = 15 (G and H), and t = 30 (I and J) show the correlated changes in Itgα5GAAKR-RFP clustering and FN-GFP aggregation in the anterior PSM. The scale bars represent 20 µm. The images are from a 94 min time-lapse, with a z stack spanning 14 µm acquired every 4 min (see Movie S2).
Reprinted from Developmental Cell, 34(1), Jülich, D., Cobb, G., Melo, A.M., McMillen, P., Lawton, A.K., Mochrie, S.G., Rhoades, E., Holley, S.A., Cross-Scale Integrin Regulation Organizes ECM and Tissue Topology, 33-44, Copyright (2015) with permission from Elsevier. Full text @ Dev. Cell