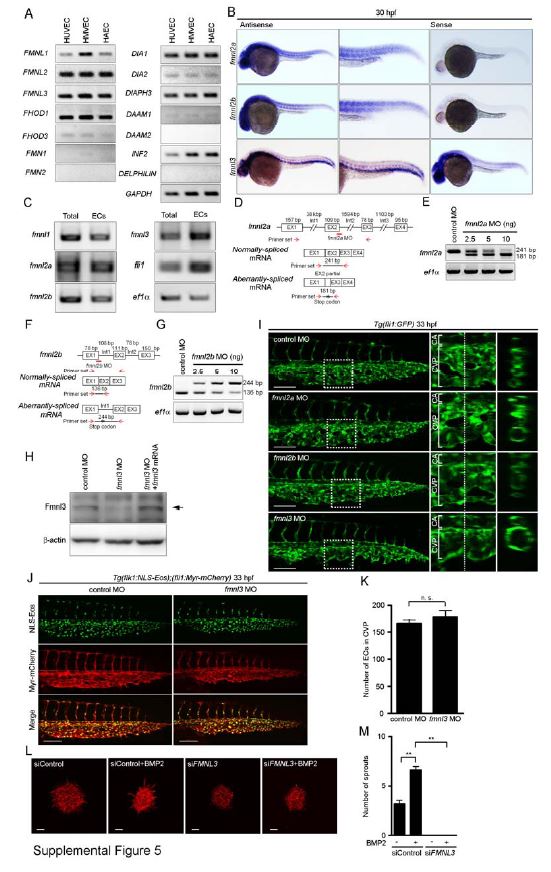
Fig. S5
Fig. S5 Fmnl3 Regulates CVP Formation. Related to Figure 5. (A) Expression of mRNAs for formin family proteins (FMNL1, FMNL2, FMNL3, FHOD1, FHOD3, FMN1, FMN2, DIA1, DIA2, DIAPH3, DAAM1, DAAM2, INF2, DELPHILIN) in the ECs (human umbilical vein EC [HUVEC], human microvascular EC [HMVEC], human arterial EC [HAEC]) was analyzed by RT-PCR analyses. (B) Expression patterns of fmnl2a, fmnl2b and fmnl3 mRNAs in zebrafish embryos at 30 hpf, as detected by whole-mount in situ hybridization. The caudal regions are enlarged at the middle column. Sense probes were used to confirm the specificity of the hybridization reactions (right column). (C) RT-PCR analyses were performed using the specific primers for the genes indicated at the left. RNAs were extracted from the 48 hpf Tg(fli1:GFP) embryos (Total) or from the FACS-sorted GFP-positive ECs (ECs). (D) fmnl2a MO (red line) is a splice-blocking MO that targets the boundary between exon 2 and intron 2. Schematic diagrams of fmnl2a gene (top), normally-spliced mRNA (middle) and aberrantly-spliced mRNA produced in fmnl2a MO-injected embryos (bottom). The size of exon/intron is indicated at the top of each exon/intron. EX, exon; Int, intron. (E) RT-PCR analyses using a primer set indicated in D were performed to assess the knockdown efficiency of fmnl2a MO similar to Figure S4B and S4C. The 241-bp fragment corresponding to normally-spliced fmnl2a mRNA was PCR-amplified in control MO-injected embryos. In contrast, PCR for fmnl2a MO-injected embryos produced the 181-bp fragment that partially lacks exon2, leading to the creation of premature stop codon indicated by asterisk. (F and G) Knockdown efficiency of fmnl2b splice-blocking MO (red line) that targets the boundary between exon 1 and intron 1 were assessed by RT-PCR analyses, as in D and E. PCR using a primer set indicated in F produced the 181-bp fragment corresponding to normally-spliced fmnl2b mRNA in control MO-injected embryos. In contrast, the 244-bp fragment that additionally contains intron 1 was PCR-amplified in fmnl2b MO-injected embryos, leading to the creation of premature stop codon as indicated by asterisk. (H) Lysates from 48 hpf zebrafish embryos injected with 2.5 ng control MO, 2.5 ng fmnl3 MO or both 2.5 ng fmnl3 MO and 100 pg MO-resistant fmnl3 mRNA were subjected to Western blot analysis with the antibody that recognizes both Fmnl2a/b and Fmnl3 and with anti-β-actin antibody. The bands corresponding to Fmnl3 are indicated by the arrow. (I) Confocal z-stack images of the caudal regions of 33 hpf Tg(fli1:GFP) embryos injected with 2.5 ng control MO, 5 ng fmnl2a MO, 5 ng fmnl2b MO or 2.5 ng fmnl3 MO are shown, as in Figure S4F. (J) Confocal z-stack images of the caudal regions of 33 hpf Tg(flk1:NLS-Eos);(fli1:Myr-mCherry) embryos injected with 2.5 ng control MO or 2.5 ng fmnl3 MO. Eos (NLS-Eos) and mCherry (Myr-mCherry) images and the merged images are shown as indicated at the left. (K) The number of ECs in the CVP as observed in J was counted, and shown as mean ± s.e.m. (control MO [n=5], fmnl3 MO [n=5]). (L) Projection view of confocal z-stack images of the spheroids formed by HUVECs transfected with control siRNA (siControl) or FMNL3 siRNA (siFMNL3), and stimulated without or with BMP2 are shown, as in Figure S3O. (M) The number of sprouts extended from each spheroid as observed in L was quantified (siControl [n=3], siControl + BMP2 [n=3], siFMNL3 [n=3], siFMNL3 + BMP2 [n=3]). Scale bars: 100 µm (I and J) and 60 µm (L). **p<0.01. n.s., no significance.
Reprinted from Developmental Cell, 32, Wakayama, Y., Fukuhara, S., Ando, K., Matsuda, M., Mochizuki, N., Cdc42 Mediates Bmp-Induced Sprouting Angiogenesis through Fmnl3-Driven Assembly of Endothelial Filopodia in Zebrafish, 109-22, Copyright (2015) with permission from Elsevier. Full text @ Dev. Cell