Fig. 2
Fig. 2
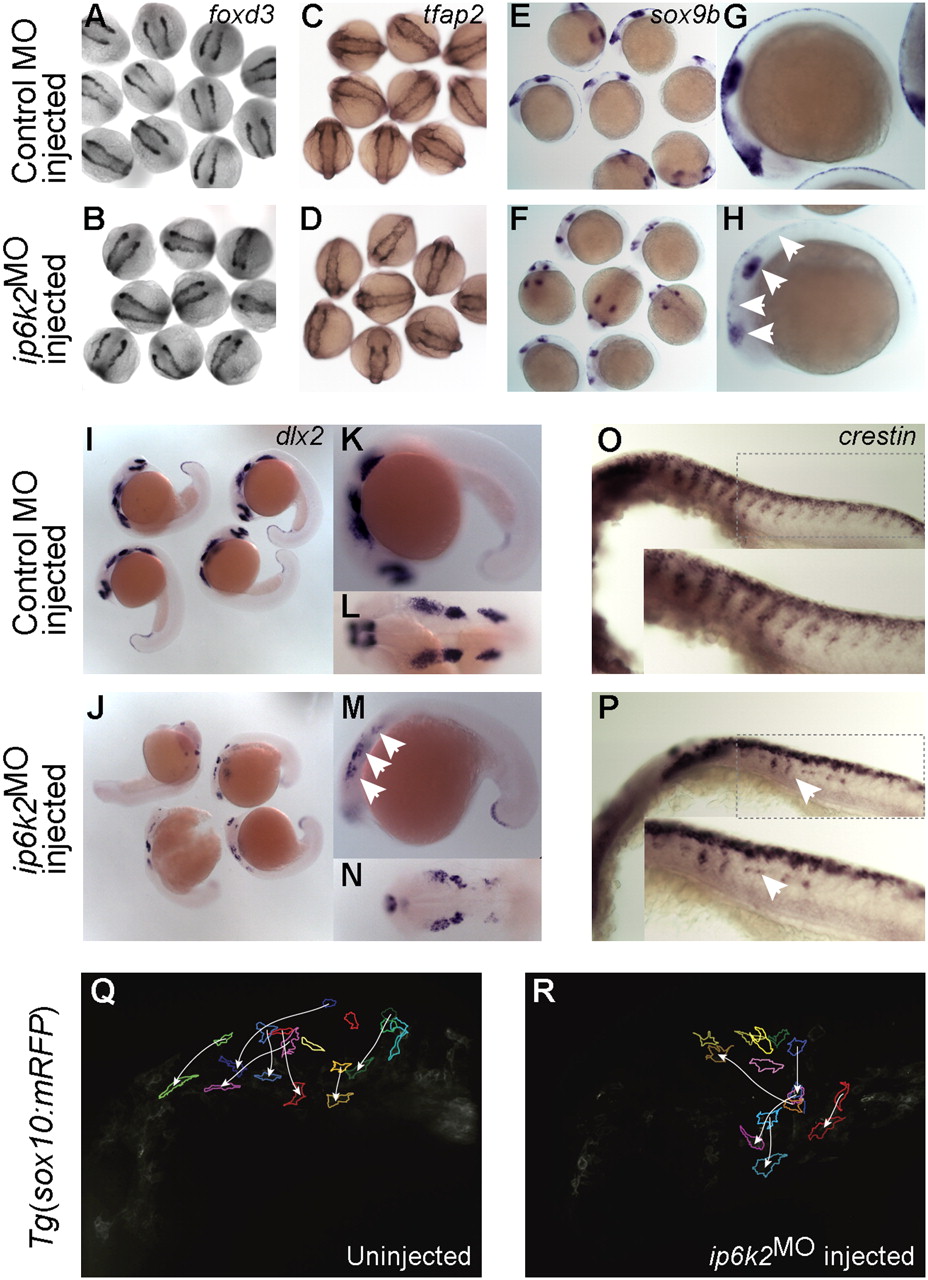
IP6K2 is critical for NCC development and migration. (A–P) Whole-mount in situ hybridizations for NC marker gene expression specifying distinct stages of NC development: (A–D) specification (foxd3 and tfap2 at 11 hpf), (E and H) maintenance (sox9b at 14 hpf), and (I–P) migration (dlx2 and crestin, 22 hpf) in control MO (A, C, E, G, I, K, L, and O) and ip6k2ATGMO (B, D, F, H, J, M, N, and P)-injected embryos. Arrowheads indicate distinct differences in the expression of sox9b (E–H), dlx2 (I–N), and crestin (O and P) between control MO and ip6k2ATGMO embryos. (Q and R) Analysis of NCC migration in real time using Tg(sox10(7.2):mrfp) fish (17) (see SI Materials and Methods for details). Fluorescence images (frames) for the hindbrain and eye regions were captured from a 6-h time-lapse sequence, with dorsal toward the top and anterior to the left. The 0-min time point represents 16-hpf stage. Images of uninjected (Q) and ip6k2ATGMO-injected (R) embryos show an overlay of first and last images onto which the tracings of migratory paths of CNCC were superimposed. Arrowheads indicate end point of each cell′s trajectory.