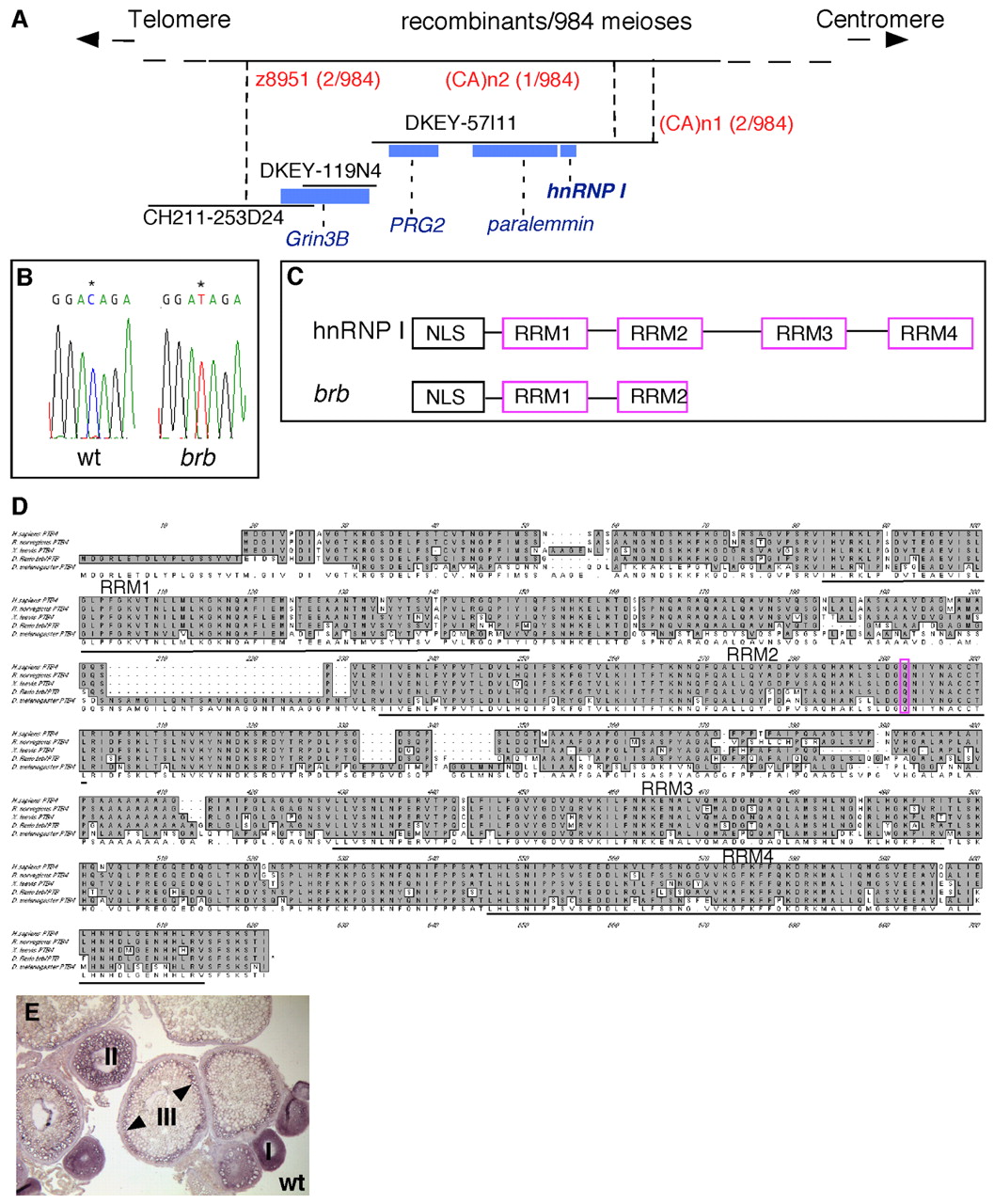
Fig. 7 Positional cloning reveals that brom bones encodes the RNA-binding protein, hnRNP I. (A) Genetic/physical map across the brom bones interval on chromosome 2. CH211-253D24, DKEY-119N4 and DKEY-57I11 are three sequenced and overlapping BAC clones in this interval. (CA)n1 and (CA)n2 are polymorphic markers we identified in BAC clone DKEY-57I11. Four transcription units were predicted based on the BAC sequence between z8951 and (CA)n2: grin3b, prg2, paralemmin and hnrnp I. Blue boxes indicate sizes and locations of these genes relative to the BAC clones. (B) Sequencing of wild-type and brom bones alleles revealed a cytosine to thymidine transition (*) in the open reading frame of the hnrnp I gene, which produces a premature stop codon. (C) Schematic diagram showing the protein domain structures of hnrnp I and brom bones. (D) Amino acid sequence alignment of hnRNP I homologs in (top to bottom) human (AAP35465), rat (Q00438), Xenopus (AAF00041), zebrafish (AAH95372) and Drosophila (NP_524703). The residue that is mutated to a stop codon in brom bones (purple) and the RRM domains are highlighted. (E) hnrnp I is expressed during oogenesis as revealed by in situ hybridization on wild-type ovary sections (arrowheads show cortical enrichment in stage III oocytes).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development