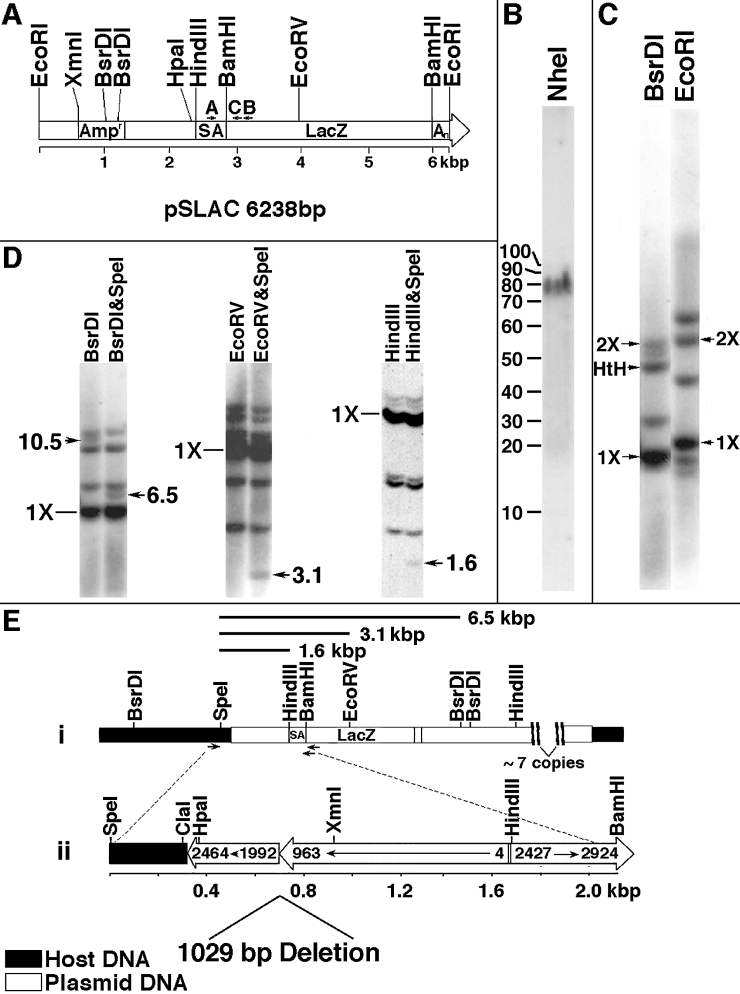
Fig. 7 Molecular characterization of the aln insertion. (A) Diagram of pSLAC linearized at the unique EcoRI site. The origins of gene sequences present in pSLAC are described under Materials and Methods. A, B, and C refer to oligomer primers used for PCR. (B) Southern blot of aln/+ genomic DNA digested with NheI, which does not cleave pSLAC. Digested DNA was subjected to FIGE and probed with radiolabeled pSLAC sequences. (C) Southern blot of aln/+ DNA digested with BsrDI or EcoRI and probed with radiolabeled pSLAC sequences. 1X refers to unit size (6.0 kbp for BsrDI, 6.2 kbp for EcoRI), 2X refers to twice unit size, and HtH refers to the predicted size of the BsrDI fragment of two units arranged head to head. (D) Identification of host–insert junction fragments. aln/+ DNA was digested with the indicated enzymes, separated on 0.7 or 1% agarose gels, blotted, and probed with radiolabeled pSLAC. Candidate junction fragments are indicated by arrows with approximate sizes given in kilobasepairs. (E) Orientation of sequences at a host–insert junction. (i) Restriction map of host and plasmid-derived sequences at one junction. Junction fragments identified in D are indicated as bars above the map. Positions of primers used to amplify junction region are indicated by arrows below the map. (ii) Diagram of the 2.1-kbp junction fragment that was PCR amplified using primers denoted in i. Host-derived sequence (324 bp) is fused to portions of two plasmid units arranged tail to tail. One plasmid unit has an internal deletion of about 1 kbp, and the other plasmid unit has a terminal deletion of about 2.4 kbp. Numbers within the bars refer to basepair numbers of pSLAC indicated in A.
Reprinted from Developmental Biology, 210(2), Cretekos, C.J. and Grunwald, D.J., alyron, an insertional mutation affecting early neural crest development in zebrafish, 322-338, Copyright (1999) with permission from Elsevier. Full text @ Dev. Biol.