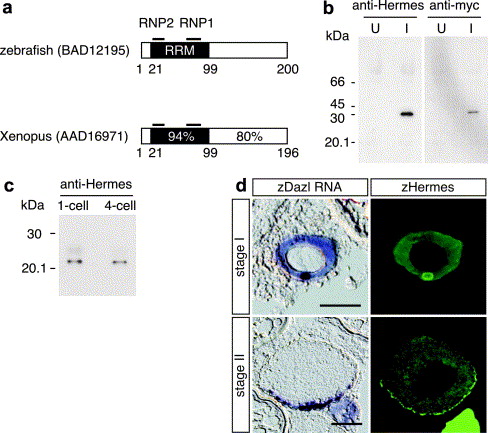
Fig. 4 Hermes protein is a constituent of the mitochondrial cloud. (a) Schematic representation of Hermes proteins in zebrafish and Xenopus (Gerber et al., 1999). RRM represents an RNA-recognition motif. The positions of conserved motifs in RRM, RNP2 and RNP1 (Birney et al., 1993) are indicated. Amino acid sequence identity among Hermes proteins in the N-terminal RRM and C-terminal regions is shown. Numbering according to amino acids in the Hermes proteins. (b) Immunodetection of Myc-tagged zebrafish Hermes using an antibody against Xenopus Hermes. The in vitro synthesized mRNA encoding Myc-zHermes was injected into 1-cell embryos and whole lysates of injected embryos were collected at 12 hpf (I). Control lysates from uninjected embryos are shown in (U). Two embryo equivalents per lane were loaded onto the gel. Immunoblotting was done with anti-Hermes and anti-Myc antibodies. As expected, the Myc-Hermes protein was detected at approximately 30 kDa. (c) Immunoblot analysis of endogenous Hermes protein in early stage zebrafish embryos (at 1-cell and 4-cell stages). (d) Distribution of dazl RNA (left) and zHermes protein (right) in the serial sections from stage I and II oocytes (upper and lower panels, respectively). Sections were cut at a thickness of 10 μm. Scale bars, 50 μm.
Reprinted from Mechanisms of Development, 124(4), Kosaka, K., Kawakami, K., Sakamoto, H., and Inoue, K., Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis, 279-289, Copyright (2007) with permission from Elsevier. Full text @ Mech. Dev.