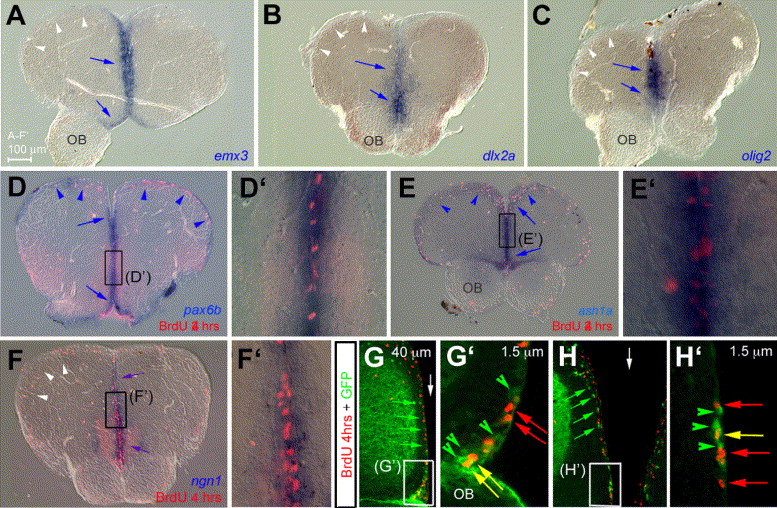
Fig. 9
Fig. 9 Unique transcription factors code and promoter usage in adult zebrafish telencephalic progenitors. (A–F′) Compared expression of emx3 (A), dlx2a (B), olig2 (C), pax6b (D, D′), ash1a (E, E′), and ngn1 (F, F′), together with BrdU incorporation after 4 h survival (red staining in panels D–F′), on anterior cross-sections (dorsal up). All markers are expressed in progenitors along the telencephalic ventricle at the midline (blue arrows, note the co-labeling with BrdU in panels D–F′). In addition, pax6b and ash1a are expressed in progenitors along the dorsally everted telencephalic ventricle (blue arrowheads in panels D, E), while other markers are not (white arrowheads in panels A–C, F). (G–H′) Determination of the enhancer fragment driving ngn1 expression in adult ventricular telencephalic progenitors by comparing BrdU incorporation (red) and GFP protein (green) in the -3.4ngn1:gfp transgenic line (Blader et al., 2004 and Blader et al., 2003). Cross-sections observed in confocal microscopy, dorsal up, green arrowheads and arrows point to cells expressing GFP only, red arrows to cells positive for BrdU only, and yellow arrows to double-labeled cells. Panels G and G2 focus on the ventral subpallium, panels H and H′ on the medial pallium, white arrows to the midline. Note that GFP distribution in -3.4ngn1:gfp is in agreement with endogenous ngn1 expression (compare with F). OB: olfactory bulb.
Reprinted from Developmental Biology, 295(1), Adolf, B., Chapouton, P., Lam, C.S., Topp, S., Tannhauser, B., Strähle, U., Gotz, M., and Bally-Cuif, L., Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon, 278-293, Copyright (2006) with permission from Elsevier. Full text @ Dev. Biol.