Fig. 1
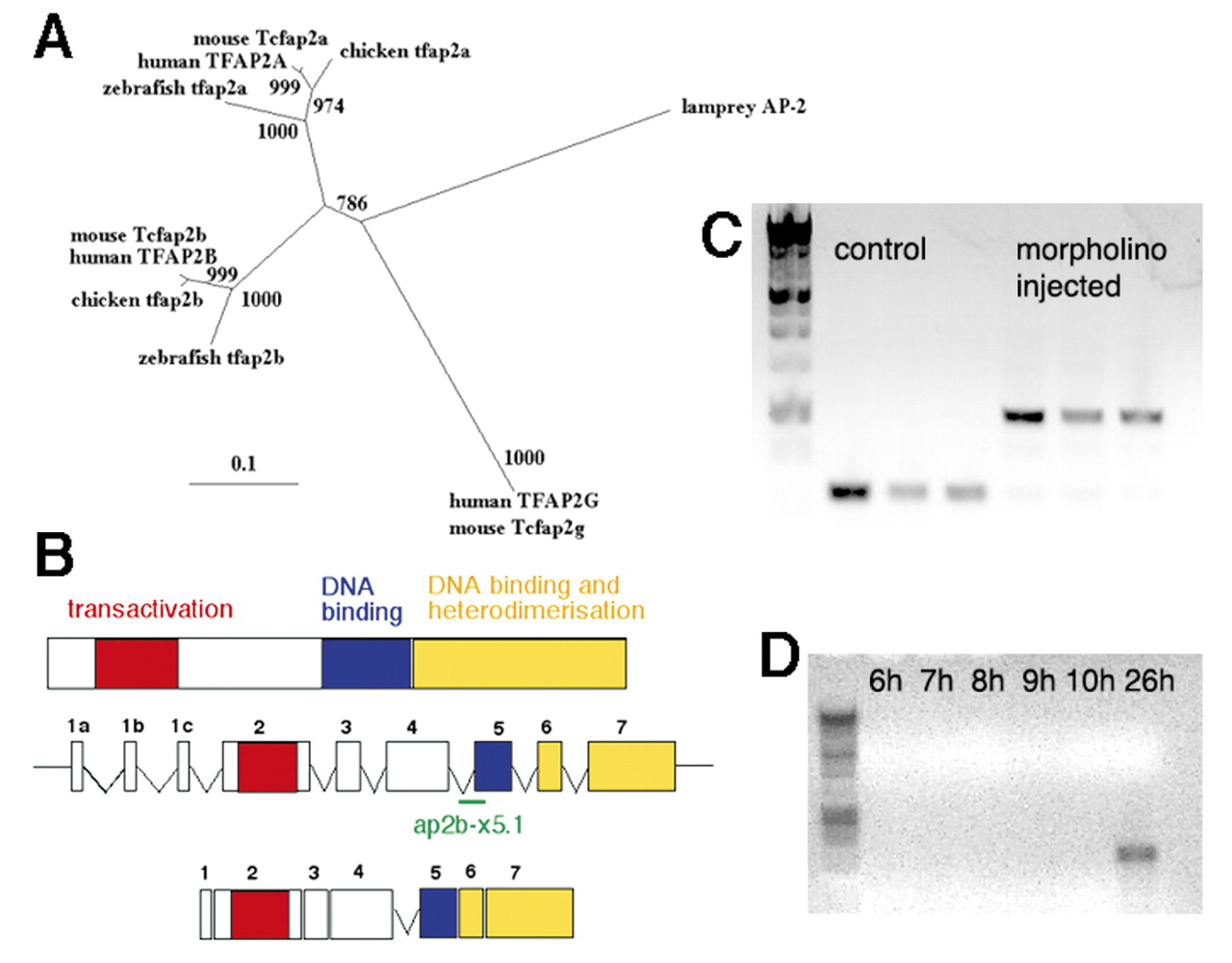
Cloning and characterization of a zebrafish tfap2b. (A) Phylogenetic tree of AP2 genes shows zebrafish tfap2b more closely related to other vertebrate AP2b genes than to other zebrafish AP2 genes. Putative protein sequences of AP2 genes were aligned using ClustalX, edited by eye and used to generate a maximum likelihood tree by quartet puzzling using the PUZZLE program. Values at the nodes are likelihood values, showing support for that node out of 1000; branch lengths represent sequence divergence. (B) tfap2b contains the highly conserved transactivation (red), DNA-binding (blue) and heterodimerization domains (yellow) found in all AP2 proteins. Exon-intron boundaries are identical in tfap2a and tfap2b, and both have three alternative first exons (1a-c). A morpholino oligonucleotide directed against the splice acceptor site before exon 5 (ap2b-x5.1-′ap2bMO′) should create a larger, unspliced product. (C) Splicing defects in tfap2b transcripts following ap2bMO injection. tfap2b was amplified from pools of ap2bMO-injected and control uninjected animals at 24 hpf using primers tfap2bf and tfap2br directed to exons 4 and 6, respectively. Uninjected animals showed PCR products of  210 bp in size in comparison with ap2bMO-injected animals in which one larger product (
210 bp in size in comparison with ap2bMO-injected animals in which one larger product ( 424 bp) was observed because of aberrant splicing. (D) PCR amplification of tfap2b from RNA isolated from different stages of zebrafish gastrulation fails to detect expression before 10 hpf
424 bp) was observed because of aberrant splicing. (D) PCR amplification of tfap2b from RNA isolated from different stages of zebrafish gastrulation fails to detect expression before 10 hpf
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development