Fig. 6
- ID
- ZDB-FIG-250423-26
- Publication
- Korzeniwsky et al., 2025 - Dominant Negative Mitf Allele Impacts Melanophore and Xanthophore Development and Reveals Collaborative Interactions With Tfec in Zebrafish Chromatophore Lineages
- Other Figures
- All Figure Page
- Back to All Figure Page
|
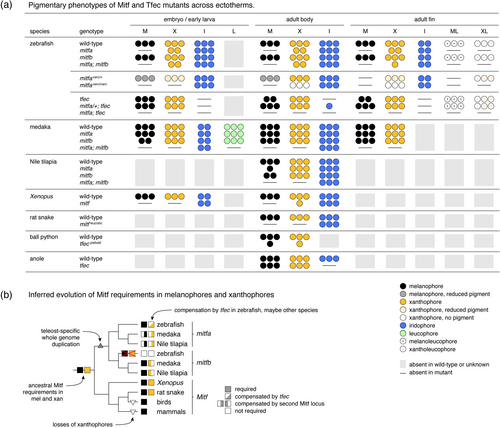
Pigmentary phenotypes of Mitf and Tfec mutants and inferred evolution of Mitf requirements. (a) Several studies have now examined phenotypes resulting from Mitf or Tfec mutation. Summary illustrates for indicated pigment cell classes absences, reduced numbers or changes in states across life stages and anatomical locations when relevant and described (this study; Garcia- Elfring et al. 2023; Lane and Lister 2012; Lister et al. 1999; Miyadai et al. 2023; Ran et al. 2024; Tzika 2024; Ullate-Agote and Tzika 2021; Wang et al. 2021). Reduced numbers of cells indicated by fewer symbols are not intended to be quantitative. Key shown at lower right. (b) Observations available so far suggest an ancestral vertebrate requirement for Mitf in both melanophores and xanthophores. With an additional whole-genome duplication in teleosts, Mitf continued to be required but loss of one Mitf locus could be compensated by another. Only in zebrafish has one Mitf locus, mitfb, undergone a substantial, though not complete (Figure 5b) reduction in pigmentary function (loss of requirements by melanophores and xanthophores marked with red X), with compensation for mitfa loss from the more distant MiT subfamily member gene, tfec, and only to a lesser extent mitfb. |