Figure 1—figure supplement 2.
- ID
- ZDB-FIG-230916-162
- Publication
- Sahai-Hernandez et al., 2023 - Dermomyotome-derived endothelial cells migrate to the dorsal aorta to support hematopoietic stem cell emergence
- Other Figures
-
- Figure 1
- Figure 1—figure supplement 1.
- Figure 1—figure supplement 2.
- Figure 1—figure supplement 3.
- Figure 2
- Figure 2—figure supplement 1.
- Figure 3
- Figure 4
- Figure 5
- Figure 5—figure supplement 1.
- Figure 6
- Figure 6—figure supplement 1.
- Figure 7.
- Figure 8—figure supplement 1.
- Figure 8—figure supplement 1.
- Figure 9.
- All Figure Page
- Back to All Figure Page
|
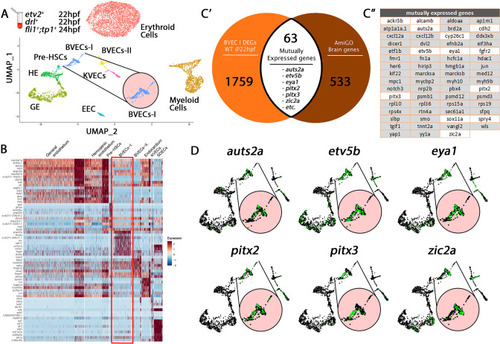
Comparison of BVECs-I cluster genes to brain annotated genes validates cluster origin. ( |