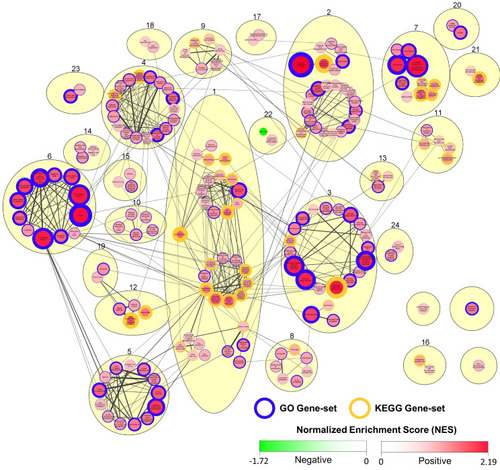
Summary of GSEA output indicating gene-set enrichment based on gene expression in CyHV-3-infected relative to mock-infected zebrafish larvae at 2 dpi. Cytoscape Network representing functional relationships between all significantly enriched gene-sets (positive or negative) identified in GSEA output (FDR adjusted p-value < 0.25). Nodes in the network represent GO (blue border) and KEGG Pathway (gold border) gene-sets. Edges (connecting lines) between nodes represent the similarity coefficient (measuring the functional/gene overlap between pairs of gene-sets). Edge thickness corresponds to magnitude of similarity coefficient (only edges with coefficient ≥2 are displayed). Each gene set exhibits either a positive or negative normalized enrichment score (NES), indicating predominant upregulation or downregulation of constituent genes, respectively. Accordingly, node colour and size both represent NES magnitude (exponentially transformed scale), with positive and negative enrichment represented by red and green, respectively, according to the colour scale provided. The node border thickness indicates the significance of enrichment (inverse of FDR adjusted p-values, thus the lower the FDR adjusted p-value, the greater the thickness). Using the MCL cluster algorithm, GO and KEGG gene-sets were clustered together based on their functional similarity as indicated by similarity coefficients (beige ovals), and numbers were assigned to each cluster. For the purposes of visual clarity, clusters were manually repositioned, and within some clusters, sub-clusters were manually grouped based on functional similarity. Clusters that are overlapping or touching in the absence of any visible edges between their respective nodes have shared edges below the 0.2 coefficient cut-off for display. Clusters that do not exhibit edges between their respective nodes and are also not touching or overlapping either have no common edges or have common edges with similarity coefficient >0.1. Higher quality figures for the whole manuscript are available in the PDF version.
|

