Fig. 2
- ID
- ZDB-FIG-221222-33
- Publication
- Daane et al., 2021 - Modulation of bioelectric cues in the evolution of flying fishes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
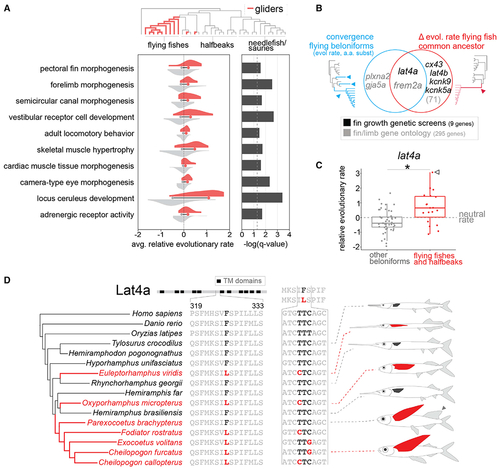
Convergent and ancestral genetic signatures in fin and limb genes in the evolution of gliding beloniforms
(A) Comparison of average relative evolutionary rate between the gliding and non-gliding beloniform nodes across specific gene ontology terms across the beloniform phylogeny (gliding fish lineages highlighted in red). Histograms represent the distribution across each node in the phylogeny of the average relative evolutionary rate for all genes within a given gene ontology term. The mean of the average relative evolutionary rates in gliding and non-gliding beloniform nodes is denoted at the base of each histogram. Significance (–log(q-value)) based on false discovery rate (FDR)-corrected p values from a Wilcoxon signed-rank test of differences between the mean relative evolutionary rate of gliding beloniforms compared to non-gliding beloniform branches. For full enrichment data, see Data S1F. (B) Intersection of molecular convergence and changes to evolutionary rate in key fin-associated genes, as defined from genetic screens and gene ontology sets, at key nodes in the Beloniformes phylogeny associated with flight. See Data S1H for full gene list. We detected parallel amino acid substitutions (Data S1G) and convergent changes to relative evolutionary rate (Data S1I) in gliding beloniforms. We further identified sequence elements under accelerated or constrained evolution in the ancestral flying fish lineage (Exocoetidae; Data S1J). (C) Elevated relative evolutionary rate in lat4a in gliding beloniforms. Each dot represents a node in the beloniform phylogeny. *p < 0.05 for two-tailed t test. White arrowhead indicates Parexocoetus brachypterus. Lat4a also has a phyloP acceleration p = 0.008 on the common ancestral node of flying fishes (Data S1J). (D) Convergent amino acid substitution in a highly conserved region of Lat4a through multiple codon mutations in flying fishes and flying halfbeaks. This substitution is found in both lineages of flying halfbeak and all flying fishes (Exocoetidae) with the exception of the sailfin flying fish, P. brachypterus, which has an elongated dorsal fin (gray arrowhead). See also S1 and S2. |