Figure 2
- ID
- ZDB-FIG-211111-46
- Publication
- Ghanta et al., 2021 - 5' modifications improve potency and efficacy of DNA donors for precision genome editing
- Other Figures
- All Figure Page
- Back to All Figure Page
|
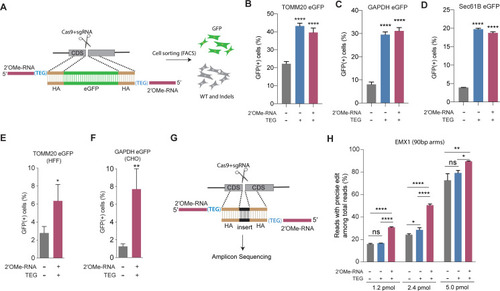
(A) Schematic representation of the 5′-modified donor design for enhanced green fluorescent protein (eGFP) insertion and fluorescence-activated cell sorting (FACS) is shown. Efficacy of eGFP integration at (B) TOMM20 and (C) GAPDH (D) Sec61B loci in HEK293T cells using unmodified, triethylene glycol (TEG) or 2′-O-methyl (2′OMe)-RNA::triethylene glycol (TEG)-modified donors are plotted as percentage of GFP+ cells. Efficacy of eGFP integration at the (E) TOMM20 locus in human foreskin fibroblast (HFF) (747 bp knock-in with ~1 kb homology arms) and (F) Gapdh locus in Chinese hamster ovary (CHO) (1635 bp knock-in with ~800 bp homology arms) cells using double-stranded DNA (dsDNA) (500 ng) donors with and without 2′OMe-RNA::TEG modifications at the 5′-ends. (G) Schematic representation of the dsDNA donor design used for quantification with deep sequencing is shown. (H) Illumina sequencing reads with precise knock-in are plotted for dsDNA donors with 90 bp homology arms at EMX1 locus in HEK293T cells. Mean ± s.d. for at least three independent replicates are plotted. p-Values were calculated using one-way ANOVA and in all cases end-modified donors were compared to unmodified donor unless indicated otherwise (Tukey’s multiple comparisons test; ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05; ns, not significant).
|